Abstract
Background
Degenerative lumbar scoliosis (DLS) progresses with aging after 50-60 years, and the genetic association of DLS remains largely unclear. In this study, the genetic association between collagen type II alpha 1 (COL2A1) gene and DLS was investigated.
Methods
COL2A1 gene polymorphism was investigated in DLS subjects compared to healthy controls to investigate the possibility of its association with COL2A1 gene. Based on a single nucleotide polymorphism (SNP) database, SNP (rs2276454) in COL2A1 were selected and genotyped using direct sequencing in 51 patients with DLS and 235 healthy controls. The SNP effects were analyzed using three models of codominant, dominant, and recessive. Logistic regression models were calculated for odds ratios (ORs) with 95% confidence intervals (CIs) and corresponding p-values, controlling age and gender as co-variables.
The adult scoliosis is defined as a de novo spinal deformity in a skeletally mature patient with a Cobb's angle greater than 10° in the coronal plain.1) Degenerative scoliosis develops after skeletal maturity without a previous history of scoliosis and typically presents in the lumbar or thoracolumbar spine.2) Degenerative lumbar scoliosis (DLS) is believed to develop as the result of asymmetric degeneration of discs, osteoporosis, and vertebral body compression fractures.3)
With the aging population of industrialized societies, lumbar spinal stenosis is an increasingly common reason for patients to seek medical attention. In addition to the spondylolisthesis, lumbar spinal stenosis can be associated with more complex spinal deformities, including variations of scoliosis.4,5)
The pathogenesis of the DLS still remains unknown, but it is presumed to be caused by the combination of genetic factors and environmental factors. Some studies performed in the oriental countries stated that long-standing squatting position can make sagittal or coronal imbalance that can result in scoliosis. There are numerous studies about idiopathic adolescent scoliosis under biochemical and genetic background,6,7,8,9) but there is no study about the genetic background on degenerative scoliosis.
There are evidences that both the collagen and glycosaminoglycans of some connective tissues in scoliosis are abnormal. The stability of the polymeric collagen in adolescents with idiopathic scoliosis was found to be significantly less than that of the control subjects.10) Furthermore, the collagen defect in the intervertebral disc might be responsible for the initiation and severity of the scoliotic curve.11)
Structural collagens are the major stress-bearing components of the spine, so the defective collagens are likely to be cause for the basic defect in scoliosis. Type II collagen is a homotrimer found in the nucleus pulposus of the intervertebral disc, and collagen type II alpha 1 (COL2A1) is encoded at the locus of type II collagen.12)
COL2A1 gene encodes the alpha-1 chain of type II collagen, a fibrillar collagen found in the cartilage and the vitreous humor of the eye. The mutation in this gene is associated with achondrogenesis type 2, Kniest dysplasia, Stickler syndrome type 1, early onset of familial osteoarthritis, spondyloepiphyseal dysplasia congenita, Langer-Saldino achondrogenesis, and Strudwick type of spondyloepimetaphyseal dysplasia.
Many authors have studied the associations of single nucleotide polymorphism (SNP) with idiopathic adolescent scoliosis, but there is no study on the association of SNP with DLS. To investigate the possibility of association of COL2A1 gene with DLS, COL2A1 gene polymorphism was investigated in subjects with DLS and healthy controls. The selected SNP was assessed based on SNP database.
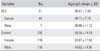
The DLS group was comprised of 51 individuals unrelated to each other (7 males and 44 females), with the mean age of 68.67 ± 7.93 years (Table 1). The DLS patients were selected from the two spine centers in Seoul, Korea. Each patient was diagnosed by two specialized spine surgeons, and all patients fulfilled the physical examination and radiographic criteria (Cobb's angle > 10°). Informed consent was obtained from all individuals, according to the Declaration of Helsinki guidelines. The study was approved by the Ethics Review Committee of the Medical Research Institute and the National Medical Center (Seoul, Korea). The average Cobb's angle of DLS group was 19.36° ± 7.38° (range, 10° to 41°), and average lateral listhesis was 4.84 ± 4.24 mm (range, 0 to 14 mm) (Table 2). The apex of the curve was located at L1 in 4 patients, L2 in 13 patients, L2-3 in 4 patients, L3 in 19 patients, L3-4 in 5 patients, and L4 in 6 patients. Among the DLS group, 32 patients had left convex curve, and 19 patients had right convex curve (Table 3).
The control group was comprised of 235 individuals (116 males and 119 females), with mean age of 59.55 ± 14.14 years (mean ± SD) (Table 1). The control group individuals were recruited following a general health check-up program to confirm that they had no clinical evidence of DLS according to radiographic criteria. The ages and genders of the control group individuals were matched to those of the DLS group (Table 1).
The blood samples were collected from all subjects in ethylenediaminetetraacetic acid (EDTA) tube for DNA extraction. Genomic DNA was extracted with 100 µL whole blood using DNA isolation kit (NucleoSpin) for mammalian blood (Macherey-Nagel GmbH & Co., Düren, Germany) and stored at -20℃ before use.
Genotyping was performed by time-of-flight mass spectrometry, using the Sequenom platform. The tested specific SNPs included SNP (rs2276454) for COL2A1. The candidate SNP examined in this study was selected on the basis of a prior association to primary or secondary osteoarthritis.13,14) We selected 1 SNP within COL2A1 gene using the following criteria. (1) All SNPs selected from COL2A1 gene were expected to have variations in level of expression. It is consisted of various SNPs from the HapMap database (http://www.hapmap.org/, genome build 34). (2) Known heterozygosity and minor allele frequency (MAF) > 0.05. (3) Validation was reported. The genotyping was performed by using Illumina targeted genotyping (TG) chip array (Illumina, San Diego, CA, USA). TG chip using molecular inversion probe technology with gene chip universal microarrays provides a method that is capable of analyzing thousands of variants in a single reaction.
The genomic DNA was amplified by using the primers for each SNP in COL2A1 gene rs2276454 (sense: 5'-GTT CAG GGA GAG GTG CTG TCC-3', anti-sense: 5'-ACA GGG GAA AGG ATG TCA CTA A-3'). The PCR products were sequenced using the ABI Prism Big Dye Terminator Cycle Sequencing System (PE Applied Biosystems, Foster City, CA, USA) and were visualized on 6% polyacrylamide 6 M urea sequencing gels on an ABI Prism 377 automatic sequencer (PE Applied Biosystems). These sequence data were analyzed using SeqManII software (DNASTAR Inc., Madison, WI, USA).
Statistical analysis was performed using the SPSS ver. 17.10 (SPSS Inc., Chicago IL, USA). The chi-square (χ2) test was used to evaluate Hardy-Weinberg equilibrium between each genotype and individual. Single SNP was analyzed using the SNPAnalyzer and SNPStats (http://bioinfo.iconcologia.net/index.php). The SNP effects were analyzed using the following three models in the order listed: codominant, dominant, and recessive. Genotype frequency of polymorphism was compared between the DLS patients and the normal control, using logistic regression models. Logistic regression analysis controlling age and gender as co-variables in all three analysis models (codominant, dominant, and recessive models for rare allele) were employed to show alternative effects of the variants.
Logistic regression models were calculated for odds ratios (OR), 95% confidence intervals (CI) and corresponding p-values, controlling age and gender as co-variables. For all statistical tests, significant level was set to 0.05.
We investigated whether the COL2A1 gene polymorphism is associated with lumbar scoliosis in Korean population, through genotyping of the selected SNP. The rs2276454 (exon34) COL2A1 polymorphism, is selected among various SNPs in the COL2A1 gene. No deviation of Hardy-Weinberg equilibrium was found in genotype distributions of any polymorphisms between DLS and control. A significant difference between DLS subjects and controls was found by using the logistic regression analysis of codominant, dominant, and recessive models. Table 4 shows the results of association for DLS and control, obtained by using SNPStats.
There was significant difference in the SNP (rs2276454) of COL2A1 genes between DLS and control. For rs2276454, the C allele was more common in DLS group compared to control group, and TC genotype marginally increased the risk of DLS by 1.90-fold (codominant: p = 0.008, OR 1.90, and 95% CI 1.17 to 3.10; dominant: p = 0.001, OR 3.85, and 95% CI 1.59 to 9.29; recessive: p = 0.36, OR 1.46, and 95% CI 0.65 to 3.29) (Table 5).
The degenerative diseases of the spine, such as spondylolisthesis, lateral listhesis, spinal stenosis, and degenerative scoliosis are presented with increasing age.4,5) A complicated DLS induces severe back or leg pain with neurological deficits by compressing neural elements and causing nerve root ischemia.13) Although the precise pathogenesis of scoliotic curvature in the spine remains unclear, several studies have been undertaken to identify the molecular or genetic basis of scoliosis development and disc degeneration.14)
The specific cause of adolescent idiopathic scoliosis has not yet been established, but it has been recognized that genetic or hereditary factors could contribute to the etiology.12,15,16,17,18) Several association studies reported the possible presence of loci in a familial form of adolescent idiopathic scoliosis.7,8,9,19,20,21) There are hypotheses that the deficiency of melatonin, a hormone secreted by the pineal gland, could play a significant role in adolescent idiopathic scoliosis.21,22,23,24,25,26)
Although several studies on molecular and SNP levels have addressed spinal deformity and degeneration, no such study has yet addressed the etiology of the DLS. Considering that the etiology of DLS is unclear, the identification of the candidate genes associated with the development of DLS may potentially be used to identify individuals at risk. Therefore, the potential of COL2A1 genes being a candidate in the development of DLS was investigated through a case-control analysis in the current study. The COL2A1 gene is located on chromosome 12q13.11-13.2, and it is important for synthesizing type II collagen which is the major collagen of cartilage. The COL2A1 gene, which encodes for the most abundant protein of articular cartilage, was initially known to be associated with primary osteoarthritis in the late 80's, and since that time, there have been conflicting reports regarding this association.
There are few studies about the correlation between DLS and generic factors. The recent study has shown that the expression of DLS is supported by various, typical copy number variation-associated structural variants of the genome,27) and significant associations between NMDA receptor genes (GRIN2B) and lateral listhesis in DLS.28)
Some authors believed that the COL2A1 gene encodes for the most abundant protein of articular cartilage, so they noted a significant association between COL2A1 SNP (rs2276454) and nodal osteoarthritis in the Newfoundland (Canada) population.29)
The current study results indicate that COL2A1 is associated with DLS in the Korean population. In a case-control analysis, the SNP (rs2276454) examined in COL2A1 was significantly associated with DLS.
Although a single SNP has been found to be related to DLS, we are not claiming that this SNP is a solitary genetic factor associated with DLS. The significance of this paper lies in the fact that it is the first study that revealed the relationship between an SNP and DLS. Additional studies are warranted, but the genetic factor should be considered as one of the multiple factors related to DLS.
This study has a number of potential limitations. The sample size is small, and the sample is limited to the Korean population. There is a need to compare our results with other ethnic groups, and also to conduct further studies about DLS and functional proteins. Thus, we expect much more researches to be developed.
In conclusion, the results of the present study revealed that we cannot deny the significance of SNP in the COL2A1 gene, as one of the multiple factors of DLS. To our knowledge, this is the first report to conduct a genetic study on DLS. Since no other studies concerning the DLS have yet been reported, more studies will be needed to confirm the possible role of SNP in the development of DLS.
Figures and Tables
References
1. Benner B, Ehni G. Degenerative lumbar scoliosis. Spine (Phila Pa 1976). 1979; 4(6):548–552.
2. Aebi M. The adult scoliosis. Eur Spine J. 2005; 14(10):925–948.
3. Herkowitz HN, Kurz LT. Degenerative lumbar spondylolisthesis with spinal stenosis: a prospective study comparing decompression with decompression and intertransverse process arthrodesis. J Bone Joint Surg Am. 1991; 73(6):802–808.
4. Tribus CB. Degenerative lumbar scoliosis: evaluation and management. J Am Acad Orthop Surg. 2003; 11(3):174–183.
5. Ploumis A, Transfledt EE, Denis F. Degenerative lumbar scoliosis associated with spinal stenosis. Spine J. 2007; 7(4):428–436.
6. Ogilvie JW, Braun J, Argyle V, Nelson L, Meade M, Ward K. The search for idiopathic scoliosis genes. Spine (Phila Pa 1976). 2006; 31(6):679–681.
7. Wise CA, Barnes R, Gillum J, Herring JA, Bowcock AM, Lovett M. Localization of susceptibility to familial idiopathic scoliosis. Spine (Phila Pa 1976). 2000; 25(18):2372–2380.
8. Salehi LB, Mangino M, De Serio S, et al. Assignment of a locus for autosomal dominant idiopathic scoliosis (IS) to human chromosome 17p11. Hum Genet. 2002; 111(4-5):401–404.
9. Chan V, Fong GC, Luk KD, et al. A genetic locus for adolescent idiopathic scoliosis linked to chromosome 19p13.3. Am J Hum Genet. 2002; 71(2):401–406.
10. Bushell GR, Ghosh P, Taylor TK, Sutherland JM. The collagen of the intervertebral disc in adolescent idiopathic scoliosis. J Bone Joint Surg Br. 1979; 61(4):501–508.
11. Wise CA, Gao X, Shoemaker S, Gordon D, Herring JA. Understanding genetic factors in idiopathic scoliosis, a complex disease of childhood. Curr Genomics. 2008; 9(1):51–59.
12. Carr AJ, Ogilvie DJ, Wordsworth BP, Priestly LM, Smith R, Sykes B. Segregation of structural collagen genes in adolescent idiopathic scoliosis. Clin Orthop Relat Res. 1992; (274):305–310.
13. Ward LM, Lalic L, Roughley PJ, Glorieux FH. Thirty-three novel COL1A1 and COL1A2 mutations in patients with osteogenesis imperfecta types I-IV. Hum Mutat. 2001; 17(5):434.
14. Arbit E, Pannullo S. Lumbar stenosis: a clinical review. Clin Orthop Relat Res. 2001; (384):137–143.
15. Cowell HR, Hall JN, MacEwen GD. Genetic aspects of idiopathic scoliosis: a Nicholas Andry Award essay, 1970. Clin Orthop Relat Res. 1972; 86:121–131.
16. Harrington PR. The etiology of idiopathic scoliosis. Clin Orthop Relat Res. 1977; (126):17–25.
17. Winter RB. Evolution in the treatment of idiopathic scoliosis in Minnesota: a family report. Minn Med. 1982; 65(10):627–629.
18. Justice CM, Miller NH, Marosy B, Zhang J, Wilson AF. Familial idiopathic scoliosis: evidence of an X-linked susceptibility locus. Spine (Phila Pa 1976). 2003; 28(6):589–594.
19. Alden KJ, Marosy B, Nzegwu N, Justice CM, Wilson AF, Miller NH. Idiopathic scoliosis: identification of candidate regions on chromosome 19p13. Spine (Phila Pa 1976). 2006; 31(16):1815–1819.
20. Gao X, Gordon D, Zhang D, et al. CHD7 gene polymorphisms are associated with susceptibility to idiopathic scoliosis. Am J Hum Genet. 2007; 80(5):957–965.
21. Miller NH, Justice CM, Marosy B, et al. Identification of candidate regions for familial idiopathic scoliosis. Spine (Phila Pa 1976). 2005; 30(10):1181–1187.
22. Machida M, Dubousset J, Imamura Y, Iwaya T, Yamada T, Kimura J. An experimental study in chickens for the pathogenesis of idiopathic scoliosis. Spine (Phila Pa 1976). 1993; 18(12):1609–1615.
23. Machida M, Dubousset J, Imamura Y, et al. Pathogenesis of idiopathic scoliosis: SEPs in chicken with experimentally induced scoliosis and in patients with idiopathic scoliosis. J Pediatr Orthop. 1994; 14(3):329–335.
24. Machida M, Dubousset J, Imamura Y, Iwaya T, Yamada T, Kimura J. Role of melatonin deficiency in the development of scoliosis in pinealectomised chickens. J Bone Joint Surg Br. 1995; 77(1):134–138.
25. Machida M, Miyashita Y, Murai I, Dubousset J, Yamada T, Kimura J. Role of serotonin for scoliotic deformity in pinealectomized chicken. Spine (Phila Pa 1976). 1997; 22(12):1297–1301.
26. Machida M, Dubousset J, Satoh T, et al. Pathologic mechanism of experimental scoliosis in pinealectomized chickens. Spine (Phila Pa 1976). 2001; 26(17):E385–E391.
27. Shin JH, Ha KY, Jung SH, Chung YJ. Genetic predisposition in degenerative lumbar scoliosis due to the copy number variation. Spine (Phila Pa 1976). 2011; 36(21):1782–1793.
28. Kim KT, Kim J, Han YJ, Kim JH, Lee JS, Chung JH. Assessment of NMDA receptor genes (GRIN2A, GRIN2B and GRIN2C) as candidate genes in the development of degenerative lumbar scoliosis. Exp Ther Med. 2013; 5(3):977–981.
29. Snelgrove TA, Peddle LJ, Stone C, et al. Association of COL1A2, COL2A1 and COL9A1 and primary osteoarthritis in a founder population. Clin Genet. 2005; 67(4):359–360.




 PDF
PDF ePub
ePub Citation
Citation Print
Print






 XML Download
XML Download