Abstract
Background and purpose
Migraine is a genetically heterogeneous disorder that is frequently associated with a familial history, and mitochondrial dysfunction has been suggested to be associated with its pathogenesis. We screened and scanned mitochondrial gene polymorphisms to determine the significance of mitochondrial DNA mutations in Korean migraineurs.
Methods
One hundred and sixty-four migraineurs aged 33.9±11.7 years (mean±SD range 12 to 65 years) were studied. Clinical data of the familial history were obtained, and blood samples were collected for DNA purification. An A-to-G substitution at mitochondrial DNA (mtDNA) position 11,084 (A11084G) was determined by a polymerase chain reaction (PCR) with BsmI restriction. In addition, new single-nucleotide polymorphism (SNP) sites in the mitochondrial genome were scanned for using PCR and direct sequencing.
Results
Ninety-eight migraine patients (59.8%) had a maternal familial history. The A11084G polymorphism, which was previously reported in 25% of Japanese migraineurs, was not evident in our Korean migraine patients. However, scanning of new SNP sites in mtDNA revealed six candidate SNPs whose incidences were higher in migraine patients than in normal subjects.
Migraine headache is a genetically heterogeneous disorder that is frequently associated with a familial history.1 Mitochondrial dysfunction may play a role in migraine pathogenesis, as proposed by the "mitochondrial hypothesis".2,3 Molecular genetic studies in migraine patients have focused on mitochondrial DNA (mtDNA) mutations that are strongly implicated in mitochondrial encephalopathies, including MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes) mutations (A3243G and T3271C), the MERRF mutation (G8344A), the Kearns-Sayre syndrome common 4977-bp deletion, and the G3460A, T4160C, and G11778A LHON mutations.4 However, most of the studies that have searched single-nucleotide polymorphisms (SNPs) or mutations in mitochondrial genomes have failed to find any candidate mutation for migraine.5
One study obtained apparently positive findings in a Japanese population. Shimomura et al. found an A-to-G substitution at mtDNA position 11,084 (A11084G) in 25% (13 out of 53) of Japanese migraineurs.6 However, a Danish study did not detect this mutation in subjects with migraine nor in subjects who had never experienced migraine.7 The Danish group concluded that the A11084G polymorphism is rare in non-Japanese migraineurs.7 However, there have been no reports of investigations undertaken to establish whether this type of substitution is present in other populations, especially in those that are ethnically similar to Japanese subjects.
Korea and Japan are geographically close, and Koreans and Japanese exhibit considerable similarity in their ethnicities.8,9 This relationship prompted us to further investigate whether the A-to-G substitution is also commonly observed in Korean migraineurs. We did not detect the A11084G polymorphism in our study population, which is consistent with the Danish result but not with the Japanese result. We therefore further scanned the entire mitochondrial genome to search for other possible candidate nucleotide polymorphic variants responsible for migraine disorder.
One hundred and sixty-four migraine patients who met the migraine criteria of ICHD-II (International Classification of Headache Disorders - second edition)10 and who visited the headache clinic at Seoul National University Hospital were enrolled. In addition, 64 control subjects without migraine who visited the neurology clinic at Seoul National University Hospital for a medical examination or routine neurological checkup, and presented with no health problems, were also included for comparison. After obtaining informed consents, detailed clinical histories including family histories and blood samples were collected. The terms "maternal inheritance" and "maternal transmission" were applied to patient whose mothers had migraine histories.
To scan the mitochondrial polymorphism in migraine patients, we enrolled a further 30 normal control subjects without history of migraine and 10 migraineurs with a maternal history. Note that we restricted the study population to patients with a maternal history due to mtDNA being transmitted maternally.
DNA was purified from whole blood using a DNA purification kit (Promega, Madison, WI). To analyze for the mitochondrial nucleotide A11084G polymorphism, mtDNA (bases 10,835 to 11,236) was amplified by the polymerase chain reaction (PCR) using previously described primers.6 The A11084G polymorphism generates a BsmI restriction site. The BsmI site containing the PCR product (bases 13,031.13,610) was also amplified as a positive control. Amplified products were digested with 1 unit of BsmI (New England Biolabs, Beverly, MA), and sized on a 1% TAE agarose gel using a 100-bp DNA ladder marker.
Fortypairs of primers were used to amplify the entire mitochondrial genome. The PCR employed 30 cycles of 96℃ for 30 seconds, 55℃ for 1 minute, and 72℃ for 30 seconds. PCR products (1 kb) were obtained for each segment, and then direct sequencing was performed. If a nucleotide sequence differed by more than 25% from the known normal sequence, it was regarded as a polymorphism. A polymorphic site was considered a candidate for migraine when its frequency was significantly higher in migraineurs than in normal controls.
Data are presented as mean±SD values and number (percentage) values, as appropriate. Frequency data between normal controls and migraineurs were compared using Pearson's chi-square test. SPSS (version 11.0 for Windows) was used for statistical analyses, with p values below 0.05 considered indicative of statistical significance.
The 164 patients with migraine comprised 123 women and 41 men aged 33.9±11.7 years (range 12 to 65 years). Family histories were present in 131 (79.9%) of the migraineurs: 98 (59.8%) in the mother, 11 (6.7%) in the father, and 22 (13.4%) in siblings and with no history of maternal or paternal transmission.
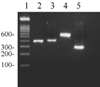
The 402-bp fragments determining the A11084G polymorphism were amplified and then cleaved by BsmI with a positive restriction control, since the A11084G polymorphism generates a BsmI restriction site. None of the 164 migraine patients showed evidence of the A11084G polymorphism, whereas all of the positive control samples were cleaved by BsmI (Fig. 1). This mutation was also not detected in the 64 normal control subjects.
We further scanned SNP sites of the mitochondrial genome from normal subjects (n=30) and migraine patients with maternal inheritance (n=10). Previous studies showed that human mtDNA comprises 16,553 bp and more than 600 polymorphic sites.11,12 In this study we identified 75 SNP sites in the 40 subjects, of which 6 exhibited a higher incidence in migraine patients than in control subjects (Table 1). T152C was present in 66.7% of the migraineurs compared to 23.8% of the normal subjects (p=0.026). The SNPs G875T (normal : migraineurs = 5.9% : 33.3%), G7599A (4.5% : 22.2%), C8793T (4.8% : 28.6%), T14667C (29.2% : 44.4%), and A15873G (4.0% : 28.6%) were also frequently detected in migraineurs, although the difference relative to controls was not statistically significant (p>0.05). Known mitochondrial mutation sites for mitochondrial disorder such as A3243G and T3271C for MELAS, and A8344G and T8356C for MERRF were not detected in either normal or patients with migraine (Fig. 2).
In this study evaluated the association between mitochondrial genes and migraine. The reported A11084G polymorphism in mtDNA in Japanese migraineurs was not observed in our Korean migraineurs, despite the close ethnicity between Japanese and Koreans. However, our negative result is consistent with that found in a Danish study.7 Although searches for mitochondrial SNPs in migraine appear to focus on the A11084G polymorphism, this is not supported by either our results or those from the Danish study.
The coding site for A11084G is the ND4 subunit of respiratory complex I, and the nucleotide substitution from A to G results in an amino acid change from threonine to alanine.13 The Japanese study found that 25% of their patients had the mutation, which was absent in 39 normal controls and 60 patients with tension-type headache.6 However, a study of 60 Danish migraineurs found no evidence of this mutation, suggesting the presence of ethnic differences.7 Subsequent studies have found incidences of the A11084G polymorphism of 14% (15/105)13 and 7.3% (35/483) in normal Japanese populations.14 Genetically distinguishing between disease-causing mutations and benign polymorphisms can be difficult due to the nonavailability of standard criteria.15 In addition, the prevalence of the target gene mutation in the patients under investigation should be known before calculating the required sample size for the healthy control subjects.15 Accordingly, the search for mtDNA SNPs in migraine needs to be reproduced in large series of studies. Nevertheless, our results suggest that the etiology of migraine in Koreans cannot be explained by the presence of the mtDNA mutation at position 11,084, and that this mutation is rare in non-Japanese populations.
Nevertheless, the genetic influence in migraine has long been discussed.1,16 In a twins study, the concordance rate of migraine was 50% in monozygotic twins but 14% in dizygotic twins, even though they were raised separately.17 The P/Q-type Ca2+-channel gene (CACNA1A) for familial hemiplegic migraine type 1,18-20 ATP1A2 or Na,K-ATPase for FHM II or III, the dopamine receptor,21-23 and the calcitonin-gene-related peptide24 have been described as being responsible for the pathogenesis of migraine. The maternal history in migraine patients, whose prevalence was also high in our study, supports the mitochondrial hypothesis of migraine, and several mitochondrial diseases including MELAS are associated with a high incidence of migraine.3 Studies employing magnetic resonance spectroscopy have revealed that mitochondrial dysfunction is present in patients with migraine, supporting the mitochondrial involvement in migraine pathophysiology.25
Accordingly, we further searched other possible candidates for mitochondrial polymorphic sites that might be account for migraine disorders. The coding proteins of T152C, G875T, G7599A, C8793T, T14667C, and A15873G are HVS2 (hypervariable segment 2), 12S ribosomal RNA, cytochrome c oxidase subunit II, ATP synthetase F0 subunit 6, NADH dehydrogenase subunit 6, and cytochrome b, respectively. Among these, ATP synthetase, NADH dehydrogenase, and cytochrome b/c might be involved in electron transport or energy generation, and dysfunction in these systems could underlie the pathogenicity of migraine.
The limitations of our study include the small number of patients, and hence the newly found polymorphic sites need to be further confirmed in studies with larger populations and including other types of headache disorders. Moreover, because we conducted our study in a tertiary general hospital, the enrolled subjects are not representative of the general migraine population, and selection bias might have been present. Accordingly, future studies are warranted that are not subject to these limitations.
Figures and Tables
Figure 1
A11084G polymorphism of mtDNA. PCR products digested with BsmI were sized on a 1% TAE agarose gel with a 100-bp DNA ladder marker (lane 1). Lane 2 is an amplified 402-bp mtDNA encompassing position 11,084. Lane 4 is a 600-bp positive control containing the BsmI site at position 13,309-13,314. Lanes 3 and 5 are PCR products digested with BsmI from lanes 2 and 4, respectively. The patterns of the PCR products from migraineurs and control subjects were identical in lanes 2 and 3.
References
3. Klopstock T, May A, Seibel P, Papagiannuli E, Diener HC, Reichmann H. Mitochondrial DNA in migraine with aura. Neurology. 1996. 46:1735–1738.

4. Sparaco M, Feleppa M, Lipton RB, Rapoport AM, Bigal ME. Mitochondrial dysfunction and migraine: evidence and hypotheses. Cephalalgia. 2006. 26:361–372.
5. Haan J, Terwindt GM, Maassen JA, Hart LM, Frants RR, Ferrari MD. Search for mitochondrial DNA mutations in migraine subgroups. Cephalalgia. 1999. 19:20–22.

6. Shimomura T KA, Marukawa H, Takahashi K. Mutation in platelet mitochondrial gene in patients with migraine. Cephalalgia. 1995. 15:10.
7. Russell MB, Diamant M, Norby S. Genetic heterogeneity of migraine with and without aura in Danes cannot be explained by mutation in mtDNA nucleotide pair 11084. Acta Neurol Scand. 1997. 96:171–173.

8. Matsumoto H, Miyazaki T, Suzuki K. Characteristics of japanese, Korean, and Chinese populations based on the genetic markers of human immunoglobulins. Z Morphol Anthropol. 1995. 81:23–32.

10. Headache Classification Subcommittee of the International Headache Society. The International Classification of Headache Disorders, 2nd edition. Cephalalgia. 2004. 24:Suppl 1. 9–160.
11. Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000. 408:708–713.

12. Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark A, Hosseini S, et al. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003. 100:171–176.

13. Sakuta R, Goto Y, Nonaka I, Horai S. An A-to-G transition at nucleotide pair 11084 in the ND4 gene may be an mtDNA polymorphism. Am J Hum Genet. 1993. 53:964–965.
14. Takeshima T, Fukuhara Y, Adachi Y, Ishizaki K, Kusumi M, Kowa H, et al. Leukocyte mitochondrial DNA A to G polymorphism at 11084 is not a risk factor for Japanese migraineurs. Cephalalgia. 2001. 21:987–989.

15. Lehmann-Horn F. Disease-causing mutations or functional polymorphisms? Acta Myol. 2004. 23:85–89.
16. Russell MB, Olesen J. Increased familial risk and evidence of genetic factor in migraine. BMJ. 1995. 311:541–544.

17. Ziegler DK, Hur YM, Bouchard TJ Jr, Hassanein RS, Barter R. Migraine in twins raised together and apart. Headache. 1998. 38:417–422.

18. Joutel A, Bousser MG, Biousse V, Labauge P, Chabriat H, Nibbio A, et al. A gene for familial hemiplegic migraine maps to chromosome 19. Nat Genet. 1993. 5:40–45.

19. Ophoff RA, Terwindt GM, Vergouwe MN, van Eijk R, Oefner PJ, Hoffman SM, et al. Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell. 1996. 87:543–552.

20. Tournier-Lasserve E. Sandler M, Ferrari M, Harnett S, editors. Genetics of familial hemiplegic migraine. Migraine: Pharmacology and Genetics. 1996. 1st edn. London: Chapman & Hall;282–290.
21. Dichgans M, Forderreuther S, Deiterich M, Pfaffenrath V, Gasser T. The D2 receptor NcoI allele: absence of allelic association with migraine with aura. Neurology. 1998. 51:928.
22. Peroutka SJ, Wilhoit T, Jones K. Clinical susceptibility to migraine with aura is modified by dopamine D2 receptor (DRD2) NcoI alleles. Neurology. 1997. 49:201–206.

23. Peroutka SJ, Price SC, Wilhoit TL, Jones KW. Comorbid migraine with aura, anxiety, and depression is associated with dopamine D2 receptor (DRD2) NcoI alleles. Mol Med. 1998. 4:14–21.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download