Abstract
Charcot-Marie-Tooth disease (CMT) is the most common form of inherited motor and sensory neuropathy. Moreover, CMT is a genetically heterogeneous disorder of the peripheral nervous system, with many genes identified as CMT-causative. CMT has two usual classifications: type 1, the demyelinating form (CMT1); and type 2, the axonal form (CMT2). In addition, patients are classified as CMTX if they have an X-linked inheritance pattern and CMT4 if the inheritance pattern is autosomal recessive. A large amount of new information on the genetic causes of CMT has become available, and mutations causing it have been associated with more than 17 different genes and 25 chromosomal loci. Advances in our understanding of the molecular basis of CMT have revealed an enormous diversity in genetic mechanisms, despite a clinical entity that is relatively uniform in presentation. In addition, recent encouraging studies - shown in CMT1A animal models - concerning the therapeutic effects of certain chemicals have been published; these suggest potential therapies for the most common form of CMT, CMT1A. This review focuses on the inherited motor and sensory neuropathy subgroup for which there has been an explosion of new molecular genetic information over the past decade.
Charcot-Marie-Tooth disease (CMT) is the most common form of inherited motor and sensory neuropathy.1 CMT is genetically and clinically a heterogeneous disorder of the peripheral nervous system, and mutations of many CMT genes are known to be responsible for the development of a variety of distinct phenotypes.2 In 1886, Charcot and Marie in France and, independently, Tooth in the United Kingdom described hereditary motor and sensory neuropathies for the first time (Fig. 1).3,4 Today, the classification of CMT has been revised and extended based on clinical features and electrophysiological, histopathological, and genetic findings.5
Autosomal-dominant inherited CMT is usually classified as type 1, the demyelinating form (CMT1), or type 2, the axonal form (CMT2).6 CMT1 patients have severely reduced nerve conduction velocities (NCVs). The upper limit of NCV for the motor median nerve in CMT1 patients is 38 m/s.7 Histopathological examinations of peripheral nerve biopsies frequently reveal extensive segmental demyelination and remyelination.8 The primary defect of CMT2 patients is neuronal.9 We classify patients as having CMTX if they have an X-linked inheritance pattern, and CMT4 if the inheritance pattern is autosomal recessive.9
Considerable new information about the pathophysiology and causative genes of CMT has recently become available.2 To date, mutations causing inherited neuropathies have been associated with at least 17 different genes, and chromosomal loci have been identified in more than 25 others (see Table 1).6 These investigations are helpful not only for the pathophysiologic studies of peripheral neuropathies but also the clinical and genetic classification of complex peripheral nerve disease. Onapristone, ascorbic acid, and neurotrophin-3 (NT3) have recently been introduced for the treatment of CMT.10-12 In this review, we focus on the 17 gene mutations that are known to cause CMT, and discuss the biological mechanisms of the peripheral neuropathies associated with myelin and axons.
Myelin plays an important role in the saltatory transmission of impulses along neuronal extensions.8 As part of the process of myelination, myelin-forming Schwann cells trap large-caliber axons within their plasma membranes during the development of the peripheral nervous system.13 Faulty communication between Schwann cells and neurons, due to genetic defects, frequently leads to these peripheral neuropathies.8
The peripheral myelin protein 22 (PMP22) gene is located within the duplication region. Several lines of evidence implicate alterations in gene dosage of PMP22 as the main factor underlying the CMT1A phenotype.14,15 This means that patients carrying one extra copy of PMP22 develop CMT1A, while patients with HNPP (hereditary neuropathy with liability to pressure palsies) deletion have only one copy of PMP22 (Fig. 2).
Tandem duplication of the CMT1A region within chromosome 17p11.2-p12, including the PMP22 gene, is the most frequent cause of CMT type 1.16 This duplication is caused by an unequal crossover event between two homologous repetitive elements flanking the 1.4-Mb region.16 Approximately 50% of all CMT patients have this duplication, as do at least 70% of all patients with CMT1.17 Patients carrying one extra copy of PMP22 develop CMT1A, whereas entire, or even partial, deletion of the 17p11.2-p12 region causes HNPP. Duplication or deletion of the 1.4-Mb fragment is the primary causative mutation, but it is not found in all cases of CMT1A and HNPP.18 In rare cases, missense or frame-shift mutations in the PMP22 gene lead to CMT1A and HNPP.19,20 The symptoms occurring in mice carrying an additional copy of PMP22 are similar to those of patients with human peripheral myelin protein duplication.21
It has been reported that the administration of the selective progesterone receptor antagonist onapristone reduces overexpression of PMP22 and impairs the CMT phenotype, without obvious side-effects.10 In addition, Passage et al. reported, with their mouse model of CMT1A, that ascorbic acid had an important effect in myelination and in reducing PMP22 concentrations to levels below those necessary to induce the disease phenotype.11
HNPP patients are characterized by recurrent pressure palsies and nerve biopsies from them show sausage-like swellings (tomacula) of the myelin sheaths.22 Deletion of the chromosome 17p11.2-p12 region that includes PMP22 frequently provides the genetic basis of hereditary peripheral demyelinating neuropathy such as HNPP (Fig. 2).23,24 Mutations and altered dosage of the PMP22 gene are regarded as the main reasons for hereditary peripheral neuropathies.25,26 Deletion is the most frequent mutation, but is not found in all cases of HNPP. In rare cases, frame-shift mutations in the PMP22 gene lead to HNPP.19,20
Clinical assessments of HNPP patients are generally less severe than those of CMT1A patients.26-28 HNPP usually develops as a painless neuropathy after minor trauma or compression. The mean ages of onset of HNPP and CMT1A are not significantly different; however, onset in the preteens was found to be more frequent in CMT1A than in HNPP.27,28
Although the point mutation of the PMP22 gene is rare, it is also a cause of CMT1A and HNPP.19,20,29 Patients carrying the PMP22 point mutation of the PMP22 gene show similar clinical features to those carrying duplication or deletion.20,29 However, in rare cases, patients with PMP22 point mutations show an early age of onset and severe phenotypes, such as Déjérine-Sottas syndrome (DSS).30 In addition, audiological evaluation can reveal auditory neuropathy in the affected individual with a frame-shift mutation Ala106fs (318delT) in the PMP22 gene.31
The early growth response 2 (EGR2) gene encodes a zinc finger transcription factor that plays a major role in myelination of peripheral nerves.32 EGR2 mutations are associated with a dominantly inherited severe form of CMT1, giving syndromes of congenital hypomyelination (CH), DSS, or CMT4E with recessive inheritance.33 Disruption of the EGR2 gene in mice blocks the development of Schwann cells at the promyelinating stage and causes demyelinating peripheral neuropathy by preventing PNS myelination.34 Thus, the normal expression of the EGR2 gene is very important in the process of Schwann cell development.34 In a recent study, overexpression of EGR2 in Schwann cells strongly increased the expression of other myelin-related genes; therefore, EGR2 mutation may cause a demyelinating form of CMT.35
A CMT family with two missense mutations in different genes has been reported.36 A R359W mutation in EGR2 was shared by the affected daughter and her father. In addition, she had a V136A mutation in GJB1, which was determined to be a de novo mutation. The daughter with two different gene mutations showed more severe clinical, electrophysiological, and histopathological phenotypes than her father, who had only the EGR2 mutation. These phenotypic differences between the proband and her father may have been caused by an altered effect of the genetic modifier in EGR2, or by the additive effect of the EGR2 and GJB1 mutations.36
The EGR2 gene regulates the expressions of myelin genes, including GJB1, PMP22, P0, and PRX in a dominant-negative manner.37,38 Mutations in EGR2 prevent Schwann cell development, and lead to the development of demyelinating neuropathy via the regulation of GJB1 expression (Fig. 3).37 Moreover, it is known that the R359W mutation in EGR2 reduces transcriptional activity in GJB1.38
Mutations in the LITAF (lipopolysaccharide-induced tumor necrosis factor-alpha factor, also referred to as SIMPLE) gene cause the demyelinating autosomal dominant disease CMT1C.9 Patients with CMT1C have decreased NCVs of 20~25 m/s, along with mild weakness and sensory loss that first presents within the first two decades of life.39 They show very similar clinical findings to CMT1A. SIMPLE is a protein encoded by the LITAF gene and may interact with Nedd4, an E3 ubiquiton ligase.40 Although SIMPLE is expressed in many cell types, when mutated it seems to cause only a demyelinating neuropathy. The disease specificity may result from the impaired degeneration of specific Shwann cell proteins.40
CMT4F is a severe from of recessive CMT that has been defined in a large Lebanese family with mutations in the periaxin (PRX) gene on chromosome 19.41 Nerve conduction is markedly slowed and onion bulbs are observed on sural nerve biopsies. PRX is a protein specifically expressed by myelinating Schwann cells.42 In adult myelinated fibers, PRX is found in the abaxonal membrane.38 During development, PRX is found in the adaxonal membrane or periaxonal cytoplasm of the myelinating Schwann cell and may have some additional function.43,44 Furthermore, an isoform of PRX is targeted to the nucleus of embryonic Schwann cells, suggesting that this protein can shuttle between the nucleus and cortical signaling or adherence complexes. Histopathological analysis of a nerve biopsy from a CMT4F patient revealed disruption of the connection between the paranodal loop and the adjacent axon; structural abnormality of the paranode was also present.45
Mutations in the gene encoding myotubularin-related phosphatase 2 (MTMR2) cause a severe autosomalrecessive, demyelinating neuropathy, CMT4B1, which has also been called hereditary motor and sensory neuropathy with focally folded myelin sheaths.46 MTMR2 contains a homologous 10-amino acid sequence with an active site for both tyrosine and serine phosphate.47 The function of MTMR2 is not yet known. However, teased fibers from sural nerve biopsy samples showed segmental demyelination associated with redundant loops of myelin, suggesting that MTMR2 has an important role in the regulation of myelin wrapping.48
Mutations in MTMR13/SBF2 have been identified in severe autosomal recessive demyelinating CMT, CMT4B2.49 SBF2 is located on chromosome 11p15 and is also known as myotubulin-related protein 13 (MTMR13). MTMR13/SBF2 is a homologue of MTMR2, which causes CMT4B1.49 This is probably the reason for CMT4B2 neuropathy resembling that of CMT4B1 both clinically and pathologically. These peripheral neuropathies show very severe disabilities in infancy and extremely slow NCVs. Sometimes patients are wheelchairbound by adulthood. Because the pathological findings of CMT4B1 and CMT4B2 are found simultaneously with the misfoldings of myelin, they probably share a common pathological mechanism. SBF2 might regulate phosphatase activity by interacting with MTMR2.9
By linkage analysis and screening genes linked to the CMT2A locus, Züchner et al. first identified several mutations in the mitofusion 2 (MFN2) gene.50 Subsequently, additional MFN2 mutations were reported in CMT2A patients (Fig. 4).51,52 Thus, mutations in MFN2 are now considered to provide the genetic basis of the CMT2A phenotype. Mitofusin 2 encodes an outer mitochondrial membrane protein which, in cooperation with the MFN1 isoform, has important roles in the regulation of mitochondrial fusion,53-55 a function essential for metabolic activity in eukaryotic cells.56 It has also been suggested that MFN2 may be associated with maintaining mitochondrial membrane potentials.57 Moreover, MFN2-deficient mice die in mid-gestation and display fragmented mitochondria.54 It is also believed, from a few population-based studies, that MFN2 mutations are most common in CMT2.58 Recently, axonal CMT neuropathy with visual impairment due to optic atrophy - designated as hereditary motor and sensory neuropathy type VI (HMSN VI) - has also been shown to be caused by mutations in the MFN2 gene.59 It is well known that HMSN VI is an axonal CMT neuropathy with optic atrophy. However, the differences between CMT2A and HMSN VI with MFN2 mutations remain to be clarified. It appears that mutational loci with high frequency might exist in MFN2 at the 94th, 105th, 280th, and 364th codons.60
Ethnic population data on MFN2 mutations are limited because the relevance of the MFN2 mutation to CMT2 has been reported only recently.50 Zuchner et al. reported 7 mutations (19%) in 36 CMT2 families representing several ethnic groups; a Japanese group reported 7 mutations (9%) in 81 axonal or unclassified CMT patients,47 and an American study found 3 mutations (23%) in 13 CMT2 families.50-52 In addition, MFN2 mutations have been identified in 22% of Korean CMT2 families.60 The mutation frequency observed by the Japanese group was lower than that found in the other studies, which may be due to sampling or analytical differences or perhaps to different genetic backgrounds. Kijima et al. performed denaturing high-performance liquid chromatography (HPLC) prior to sequence analysis, a process that may have reduced the detection rate.51
Mutations in the MFN2 gene are now viewed as the primary cause of axonal autosomal-dominant CMT2A, and it has therefore been suggested that these patients should be screened for MFN2. However, clinical and electrophysiological phenotypes of CMT patients with MFN2 mutations were significantly different in earlyand late-onset groups, and optic atrophy was found only in CMT2 patients with unusually severe phenotypes with an early onset age. In addition, MFN2 mutations show variable central nervous system (CNS) involvements.
Following the mapping of CMT2A to the short arm of chromosome 1, 1p35-p36, a missense mutation was detected in KIF1B in a Japanese CMT2A family.61 However, no other mutation has been identified in KIF1B, and it is therefore believed that another gene is involved.62,63 The kinesin superfamily is responsible for microtubule-dependent transport of a variety of organelles and vesicles.
It has been reported that mutations in the ras-related protein rab-7 (RAB7) gene cause the axonal form of CMT known as CMT2B.64 RAB7 is a member of the Rab family of small G proteins, which regulate intracellular vesicle traffic. RAB7 and its effector protein, RILP, have been shown to play a role in lysosomal transport by inducing the recruitment of dynein-dynactin motors.65 Mutations in dynactin also cause axonopathy, suggesting that two separate diseases may share a common pathway.
Mutations of the small heat-shock proteins (sHSPs) are reported to cause either CMT2 or a distal hereditary motor neuropathy (dHMN).66 It is known that sHSPs are part of a protein superfamily sharing 85 amino acid residues in a C-terminal region known as the α-crystalline domain, although the function of sHSP is unclear. However, it is believed that sHSP27 is associated with protection from apoptosis and stabilization of the cytoskeleton.67 Irobi et al.66 reported that mutations in the α-crystalline domain of sHSP22 cause dHMN type II; Evgrafov et al.68 suggested that mutations in the α-crystalline and C-terminal tail of sHSP27 cause either CMT2F or another dHMN. It seems that sHSP22 and sHSP27 interact with each other, and both diseases involve common mechanisms.69,70
Glycyl-tRNA synthetase is known to cause CMT2D.71 In four families with CMT2D, mutation of Glycyl-tRNA synthetase provided the first example of an aminoacyl tRNA synthetase which was related to human genetic disease.71 Why a mutation in a tRNA synthetase should cause only a chronic neuropathy but spare other organ systems is both unknown and surprising.
Mutations in the myelin protein zero (MPZ) gene, which is located on chromosome 1q21-q22, are present in CMT1B, CMT type 2, DSS, and CH neuropathy.72,73 It is proposed that the nature and position of the MPZ mutations largely determine the axonal and demyelinating phenotypes. MPZ is highly expressed by myelinating Schwann cells and in more than half the protein in the peripheral myelin sheet. The clinical, electrophysiological, and histopathological findings appear to be heterogeneous for MPZ mutations.74 Furthermore, the same mutation can cause different degrees of disease severity in different patients.75
The protein consists of 248 amino acids and a single extracellular immunoglobulin-related domain (which mediates homophilic adhesion), a transmembrane domain, and a short basic intracellular domain.76 MPZ knock-out mice show abnormal regulation of myelin gene expression such as upregulation of MAG and PLP and downregulation of PMP22.77 The myelination regulatory mechanism of MPZ is through adhesion-mediated signal transduction.78 By this hypothesis, the in vitro mutations of MPZ are associated with decreased MPZ-mediated adhesion and these changes cause the more severe clinical features.
Neurofilament light-chain polypeptide (NEFL) is one of the most abundant cytoskeletal components of the neuron.79 The NEFL gene encoding the neurofilament plays an important role in axonal structure, including that of the extensive fibrous network in the cytoplasm of the neuron (Fig. 5).79 Mutations in the NEFL gene usually cause axonal CMT2E neuropathy, but recent studies found that NEFL mutations cause the demyelinating forms of neuropathy common to CMT1 and DSS.80
Nerve conduction studies of CMT2E patients showed that NCV is not decreased severely. These findings suggest that the pathophysiological mechanism of the neuropathy is primary axonal damage.81 A patient with NEFL gene mutation shows proximal muscle weakness, normal NCV, and reduced motor and sensory nerve action potential amplitudes, findings compatible with typical CMT2. Transgenic mice harboring loss of NEFL genes show a normal phenotype; however, NCVs are severely reduced.82,83 Mutation of the NEFL gene could be the cause of both CMT1 and CMT2 neuropathies. Population data on the mutation frequencies of the NEFL gene are very limited. The frequencies of NEFL were 4.8% in Koreans and 2.2% in Caucasian CMT patients.84,85
CMTX is the second-most-frequent form of CMT, and is caused by mutations in the gene for gap junction protein beta 1 (GJB1: connexin 32, Cx32), which maps to chromosome Xq13.86,87 CMTX patients display the distinctive criterion of an X-linked mode of inheritance; that is, an absence of man-to-man transmission and a more severe disease phenotype in affected males than in affected females of the same age.88 Motor nerve conduction is slower in CMTX males, but ranges from slightly reduced to normal in females.89 The nature of the neuropathy in CMTX remains controversial, and it could be a primary axonal neuropathy or a primary demyelinating neuropathy with secondary axonal degeneration.90,91
The GJB1 gene is involved in the transport of small molecules within Schwann cells.92 Gap junctions allow electrical communication between cells in the nervous system and GJB1 mutations affect the function of gap junctions in the myelin sheath.93 Gap junctions comprise intercellular channels among adjacent cells and they are distributed in the liver, kidneys, and CNS as well as the peripheral nerves.94 Since the GJB1 gene is expressed not only in Schwann cells but also in oligodendrocytes, GJB1 mutation has been reported to cause CNS lesions.95 There are over 200 mutations associated with various degrees of muscular weakness, atrophy, and sensory impairment. In studies of CMTX patients, mutation of GJB1 more or less destroyed the structure of the myelin and the action potential amplitude was more affected than the NCV.96 Sural nerve biopsy confirmed that there was prominent axonal loss.97
The mutation frequency of GJB1 in Korea (7.1%)85 is considerably lower than in several European groups - Spain, 21.3%98; Finland, 19.0%99; Russia, 13.0%100; Italy, 16.7%101; Germany, 11.9%102 - but is similar to that in Japan (5.6~5.7%).103,104 It appears that mutations in GJB1 are less frequent in East Asian CMT patients than in European patients.
CMT4A is linked to 8q13-q21.1 and is caused by mutations in ganglioside-induced differentiated associated protein 1 (GDAP1), a novel protein of unknown function. 105 Clinical symptoms begin at an early age (before 10 years), with delayed developmental milestones of sitting or walking; weakness spreads to proximal muscles by the end of the first decade. However, sensory loss is mild. In other patients, hoarse voice and vocal cord paresis have been reported. Axonal and demyelinating phenotypes have been associated with GDAP1 mutations, which could be the cause of both the demyelinating and axonal neuropathies.106 The GDAP1 gene is found in the nerve cell at an early stage of development but its expression is found in the Schwann cell.107 It is not yet known if mutation of the GDAP1 gene damages the nerve cell, Schwann cell, or both. However, a likely hypothesis is that mutation of the GDAP1 gene disturbs signal transmission between Schwann cells and nerve cells.
Mutations in the Gigaxonin gene lead to a rare autosomal-recessive giant axonal neuropathy (GAN) affecting the peripheral nervous system and CNS.108 Prominent axonal swelling occurs, with masses of tightly woven neurofilaments - commonly found near nodes of Ranvier - producing an increase in axon caliber.109 This causes the main stigmata of the disease. Abnormalities in the organization of neurofilaments are also found in the brains of these patients, and are the possible cause of the associated mental retardation.
GAN includes abnormal intermediate neurofilaments and displays the typically kinky-hair-like filaments. The mechanism of the disease is unknown, but the peripheral neuropathy is developed by the mutation of the Gigaxonin gene, which possibly induces changes in the axon transport system and a resulting deficiency in energy.
CMT4C is caused by homozygous or compound heterozygous mutations in the previously uncharacterized KIAA1985 gene.110 CMT4C is a childhood-onset disease associated with an early-onset scoliosis and a distinct Shwann cell pathology. Scoliosis is prominent early on and may precede weakness and sensory loss. The protein encoded by KIAA1985 belongs to new vertebrate protein group whose function is not known. By means of comparative sequence alignment, it is possible to determine that this protein group includes the multiple SH3 and TRP domains which are associated with formation of protein polymers.110
Seventeen genes that cause hereditary motor and sensory neuropathy have been described. Even though the severities of the clinical symptoms differ, CMT is caused by a single gene mutation and follows Mendel's law. Development of the disease is directly due to a genetic defect, which means that detection of the defect provides a positive diagnosis. Over the past decade, new information about the function of genes known to cause CMT and new rational treatment approaches for CMT1A have emerged.10-12 Apparent therapeutic effects of certain chemicals tested in CMT1A animal models highlight the importance of exact and speedy determination of chromosome 17p11.2-p12 duplication in subjects with peripheral neuropathies.10,11 A personalized therapy for patients with CMT1A duplication might be possible in the near future. For this reason, the importance of rapid and accurate molecular diagnosis is again emphasized. Understanding mutations and their causes and clarifying the pathophysiologic mechanisms of CMT is important not only for diagnosis but also for developing new therapies.
Figures and Tables
 | Figure 1Contemporary portraits of Charcot (A), Marie (B), and Tooth (C). In 1886, Frenchmen Jean Martin Charcot (1825~1893) and Pierre Marie (1853~1940), and Briton Howard Henry Tooth (1856~1925), described hereditary motor and sensory neuropathies for the first time. |
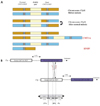 | Figure 2(A) CMT1A duplication and HNPP deletion are reciprocal products of a recombination event (unequal crossing-over) during meiosis, mediated by the flanking repeat elements (CMT1A-REPs). (B) Genomic map of the chromosome 17p11.2-p12. The genomic structure of the CMT1A/HNPP region (telomere-to-centromere orientation). Proximal and distal CMT1A-REPs are shown as vertical boxes. |
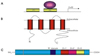 | Figure 3Genomic structures of GJB1 (A) and EGR2 (B). The EGR2 gene encodes a zinc finger transcription factor that plays a major role in myelination of the peripheral nerves, and the EGR2 gene regulates the expressions of GJB1. Thus, mutations in EGR2 prevent Schwann cell development and lead to the development of demyelinating neuropathy (C). |
 | Figure 4Genomic structure and mutations of MFN2. Solid black boxes and solid white boxes indicate protein coding sequences and untranslated sequences, respectively. P, loop; GTP, binding-site motif; Cc, coiled-coil domain; TM, transmembrane domain; GTPase, GTPase functional domain; fzo mitofusin, fzo mitofusin functional domain. |
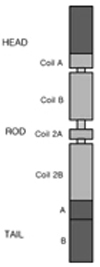 | Figure 5Structure of the NEFL protein, which is one of the most abundant cytoskeletal components of the neuron. The NEFL gene encoding the neurofilament light chain plays an important role in axonal structure, including an extensive fibrous network in the cytoplasm of the neuron. |
References
1. Dyck PJ, Chance P, Lebo R, Carney JA. Dyck PJ, Thomas PK, Griffin JW, Low PA Poduslo JF, editors. Hereditary motor and sensory neuropathies. Peripheral neuropathy. 1993. 3rd edn. Philadelphia: WB Saunders;1094–1136.
2. Berger P, Young P, Suter U. Molecular cell biology of Charcot-Marie-Tooth disease. Neurogenetics. 2002. 4:1–15.

3. Charcot J, Marie P. Sue une forme particulaire d'atrophie musculaire progressive souvent familial debutant par les pieds et les jamber et atteingnant plus tard les mains. Rev Med. 1886. 6:97–138.
4. Tooth H. The peroneal type of progressive muscular atrophy. 1886. London: Lewis.
5. Shy ME, Garbern JY, Kamholz J. Hereditary motor and sensory neuropathies: a biological perspective. Lancet Neurol. 2002. 1:110–118.

7. Harding AE, Thomas PK. The clinical features of hereditary motor and sensory neuropathy types I and II. Brain. 1980. 103:259–280.

9. Suter U, Scherer SS. Disease mechanisms in inherited neuropathies. Nat Rev Neurosci. 2003. 4:714–726.

10. Sereda MW, Meyer zu Horste G, Suter U, Uzma N, Nave KA. Therapeutic administration of progesterone antagonist in a model of Charcot-Marie-Tooth disease (CMT-1A). Nat Med. 2003. 9:1533–1537.

11. Passage E, Norreel JC, Noack-Fraissignes P, Sanguedolce V, Pizant J, Thirion X, et al. Ascorbic acid treatment corrects the phenotype of a mouse model of Charcot-Marie-Tooth disease. Nat Med. 2004. 10:396–401.

12. Sahenk Z, Nagaraja HN, McCracken BS, King WM, Freimer ML, Cedarbaum JM, et al. NT-3 promotes nerve regeneration and sensory improvement in CMT1A mouse models and in patients. Neurology. 2005. 65:681–689.

14. Huxley C, Passage E, Manson A, Putzu G, Figarella-Branger D, Pellissier JF, et al. Construction of a mouse model of Charcot-Marie-Tooth disease type 1A by pronuclear injection of human YAC DNA. Hum Mol Genet. 1996. 5:563–569.

15. Lupski JR. Charcot-Marie-Tooth disease: a gene-dosage effect. Hosp Pract (Minneap). 1997. 32:83–84. 89–91. 94–95.

16. Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, Guzzetta V, Trask BJ, et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991. 66:219–232.

17. Boerkoel CF, Takashima H, Garcia CA, Olney RK, Johnson J, Berry K, et al. Charcot-Marie-Tooth disease and related neuropathies: mutation distribution and genotype-phenotype correlation. Ann Neurol. 2002. 51:190–201.

18. Chance PF, Alderson MK, Leppig KA, Lensch MW, Matsunami N, Smith B, et al. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993. 72:143–151.

19. Young P, Wiebusch H, Strogbauer F, Ringelstein B, Assmann G, Funke H. A novel frameshift mutation in PMP22 accounts for hereditary neuropathy with liability to pressure palsies. Neurology. 1997. 48:450–452.

20. Nicholson GA, Valentijn LJ, Cherryson AK, Kennerson ML, Bragg TL, DeKroon RM, et al. A frame shift mutation in the PMP22 gene in hereditary neuropathy with liability to pressure palsies. Nat Genet. 1994. 6:263–266.

21. Sereda M, Griffiths I, Puhlhofer A, Stewart H, Rossner MJ, Zimmerman F, et al. A transgenic rat model of Charcot-Marie-Tooth disease. Neuron. 1996. 16:1049–1060.

22. Behse F, Buchthal F, Carlsen F, Knappeis GG. Hereditary neuropathy with liability to pressure palsies. Electrophysiological and histopathological aspects. Brain. 1972. 95:777–794.

23. Verhagen WI, Gabreels-Festen AA, van Wensen PJ, Joosten EM, Vingerhoets HM, Gabreels FJ, et al. Hereditary neuropathy with liability to pressure palsies: a clinical, electroneurophysiological and morphological study. J Neurol Sci. 1993. 116:176–184.

24. Le Guern E, Sturtz F, Gugenheim M, Gouider R, Bonnebouche C, Ravise N, et al. Detection of deletion within 17p11.2 in 7 French families with hereditary neuropathy with liability to pressure palsies (HNPP). Cytogenet Cell Genet. 1994. 65:261–264.

25. Mariman FC, Sillekens PT, van Beek-Reinders, van Venrooij WJ. A model for the excision of introns 1 and 2 from adenoviral major late pre-messenger RNAs. J Mol Biology. 1984. 178:47–62.

26. Gouider R, LeGuern E, Emile J, Tardieu S, Cabon F, Samid M, et al. Hereditary neuralgic amyotrophy and hereditary neuropathy with liability to pressure palsies: two distinct clinical, electrophysiologic, and genetic entities. Neurology. 1994. 44:2250–2252.

27. Pareyson D, Scaioli V, Taroni F, Botti S, Lorenzetti D, Solari A, et al. Phenotypic heterogeneity in hereditary neuropathy with liability to pressure palsies associated with chromosome 17p11.2-12 deletion. Neurology. 1996. 46:1133–1137.

28. Kim SM, Chung KW, Choi BO, Yoon ES, Choi JY, Park KD, et al. Hereditary neuropathy with liability to pressure palsies (HNPP) patients of Korean ancestry with chromosome 17p11.2-p12 deletion. Exp Mol Med. 2004. 36:28–35.

29. Roa BB, Garcia CA, Suter U, Kulpa DA, Wise CA, Mueller J, et al. Charcot-Marie-Tooth disease type 1A. Association with a spontaneous point mutation in the PMP22 gene. N Engl J Med. 1993. 329:96–101.

30. Roa BB, Dyck PJ, Marks HG, Chance PF, Lupski JR. Déjérine-Sottas syndrome associated with point mutation in the peripheral myelin protein 22 (PMP22) gene. Nat Genet. 1993. 5:269–273.

31. Choi BO, Chung KW, Park KD, Choi KG, Kim SM, Kim Y, et al. Charcot-Marie-Tooth type 1A patient with a novel frame shift mutation (Ala106fs) in the PMP22 gene. J Korean Neurol Assoc. 2004. 22:673–676.
32. Timmerman V, De Jonghe P, Ceuterick C, De Vriendt E, Lofgren A, Nelis E, et al. Novel missense mutation in the early growth response 2 gene associated with Dejerine-Sottas syndrome phenotype. Neurology. 1999. 52:1827–1832.

33. Warner LE, Mancias P, Butler IJ, McDonald CM, Keppen L, Koob KG, et al. Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat Genet. 1998. 18:382–384.

34. Topilko P, Schneider-Maunoury S, Levi G, Baron-Van Evercooren A, Chennoufi AB, Seitanidou T, et al. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994. 371:796–799.

35. Nagarajan R, Svaren J, Le N, Araki T, Watson M, Milbrandt J. EGR2 mutations in inherited neuropathies dominant-negatively inhibit myelin gene expression. Neuron. 2001. 30:355–368.

36. Chung KW, Sunwoo IN, Kim SM, Park KD, Kim WK, Kim TS, et al. Two missense mutations of EGR2 R359W and GJB1 V136A in a Charcot-Marie-Tooth disease family. Neurogenetics. 2005. 6:159–163.

37. Musso M, Balestra P, Bellone E, Cassandrini D, Di Maria E, Doria LL, et al. The D355V mutation decreases EGR2 binding to an element within the Cx32 promoter. Neurobiol Dis. 2001. 8:700–706.

38. Warner LE, Svaren J, Milbrandt J, Lupski JR. Functional consequences of mutations in the early growth response 2 gene (EGR2) correlate with severity of human myelinopathies. Hum Mol Genet. 1999. 8:1245–1251.

39. Tang X, Fenton MJ, Amar S. Identification and functional characterization of a novel binding site on TNF-alpha promoter. Proc Natl Acad Sci U S A. 2003. 100:4096–4101.

40. Moriwaki Y, Begum NA, Kobayashi M, Matsumoto M, Toyoshima K, Seya T. Mycobacterium bovis Bacillus Calmette-Guerin and its cell wall complex induce a novel lysosomal membrane protein, SIMPLE, that bridges the missing link between lipopolysaccharide and p53-inducible gene, LITAF (PIG7), and estrogen-inducible gene, EET-1. J Biol Chem. 2001. 276:23065–23076.

41. Guilbot A, Williams A, Ravise N, Verny C, Brice A, Sherman DL, et al. A mutation in periaxin is responsible for CMT4F, an autosomal recessive form of Charcot-Marie-Tooth disease. Hum Mol Genet. 2001. 10:415–421.

42. Gillespie CS, Sherman DL, Blair GE, Brophy PJ. Periaxin, a novel protein of myelinating Schwann cells with a possible role in axonal ensheathment. Neuron. 1994. 12:497–508.

43. Sherman DL, Brophy PJ. A tripartite nuclear localization signal in the PDZ-domain protein L-periaxin. J Biol Chem. 2000. 275:4537–4540.

44. Scherer SS, Xu YT, Bannerman PG, Sherman DL, Brophy PJ. Periaxin expression in myelinating Schwann cells: modulation by axon-glial interactions and polarized localization during development. Development. 1995. 121:4265–4273.

45. Kijima K, Numakura C, Shirahata E, Sawaishi Y, Shimohata M, Igarashi S, et al. Periaxin mutation causes early-onset but slow-progressive Charcot-Marie-Tooth disease. J Hum Genet. 2004. 49:376–379.

46. Bolino A, Muglia M, Conforti FL, LeGuern E, Salih MA, Georgiou DM, et al. Charcot-Marie-Tooth type 4B is caused by mutations in the gene encoding myotubularinrelated protein-2. Nat Genet. 2000. 25:17–19.

47. Laporte J, Blondeau F, Buj-Bello A, Mandel JL. The myotubularin family: from genetic disease to phosphoinositide metabolism. Trends Genet. 2001. 17:221–228.

48. Gambardella A, Bono F, Muglia M, Valentino P, Quattrone A. Autosomal recessive hereditary motor and sensory neuropathy with focally folded myelin sheaths (CMT4B). Ann NY Acad Sci. 1999. 883:47–55.

49. Senderek J, Bergmann C, Weber S, Ketelsen UP, Schorle H, Rudnik-Schoneborn S, et al. Mutation of the SBF2 gene, encoding a novel member of the myotubularin family, in Charcot-Marie-Tooth neuropathy type 4B2/11p15. Hum Mol Genet. 2003. 12:349–356.

50. Züchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004. 36:449–451.

51. Kijima K, Numakura C, Izumino H, Umetsu K, Nezu A, Shiiki T, et al. Mitochondrial GTPase mitofusin 2 mutation in Charcot-Marie-Tooth neuropathy type 2A. Hum Genet. 2005. 116:23–27.

52. Lawson VH, Graham BV, Flanigan KM. Clinical and electrophysiologic features of CMT2A with mutations in the mitofusin 2 gene. Neurology. 2005. 65:197–204.

53. Rojo M, Legros F, Chateau D, Lombes A. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J Cell Sci. 2002. 115:1663–1674.

54. Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003. 160:189–200.

55. Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004. 305:858–862.

56. Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005. 280:26185–26192.

57. Honda S, Aihara T, Hontani M, Okubo K, Hirose S. Mutational analysis of action of mitochondrial fusion factor mitofusin-2. J Cell Sci. 2005. 118:3153–3161.

58. Szigeti K, Garcia CA, Lupski JR. Charcot-Marie-Tooth disease and related hereditary polyneuropathies: molecular diagnostics determine aspects of medical management. Genet Med. 2006. 8:86–92.

59. Züchner S, De Jonghe P, Jordanova A, Claeys KG, Guergueltcheva V, Cherninkova S, et al. Axonal neuropathy with optic atrophy is caused by mutations in mitofusin 2. Ann Neurol. 2006. 59:276–281.

60. Choi BO, Kim SB, Park KD, Choi KG, Oh J, Suh BC, et al. Clinical and genetic characteristics in patients of Charcot-Marie-Tooth type 2A with mitofusin 2 (MFN2) mutations. J Korean Neurol Assoc. 2006. 24:131–140.
61. Zhao C, Takita J, Tanaka Y, Setou M, Nakagawa T, Takeda S, et al. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1B beta. Cell. 2001. 105:587–597.

62. Bissar-Tadmouri N, Nelis E, Züchner S, Parman Y, Deymeer F, Serdaroglu P, et al. Absence of KIF1B mutation in a large Turkish CMT2A family suggests involvement of a second gene. Neurology. 2004. 62:1522–1525.

63. Muglia M, Zappia M, Timmerman V, Valentino P, Gabriele AL, Conforti FL, et al. Clinical and genetic study of a large Charcot-Marie-Tooth type 2A family from southern Italy. Neurology. 2001. 56:100–103.

64. Verhoeven K, De Jonghe P, Coen K, Verpoorten N, Auer-Grumbach M, Kwon JM, et al. Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy. Am J Hum Genet. 2003. 72:722–727.

65. Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol. 2001. 11:1680–1685.

66. Irobi J, Van Impe K, Seeman P, Jordanova A, Dierick I, Verpoorten N, et al. Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nat Genet. 2004. 36:597–601.

68. Evgrafov OV, Mersiyanova I, Irobi J, Van Den Bosch L, Dierick I, Leung CL, et al. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet. 2004. 36:602–606.

69. Sun X, Fontaine JM, Rest JS, Shelden FA, Welsh MJ, Benndorf R. Interaction of human HSP22 (HSPB8) with other small heat shock proteins. J Biol Chem. 2004. 279:2394–2402.

70. Benndorf R, Sun X, Gilmont RR, Biederman KJ, Molloy MP, Goodmurphy CW, et al. HSP22, a new member of the small heat shock protein superfamily, interacts with mimic of phosphorylated HSP27 ((3D)HSP27). J Biol Chem. 2001. 276:26753–26761.

71. Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin SQ, et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet. 2003. 72:1293–1299.

72. Warner LE, Hilz MJ, Appel SH, Killian JM, Kolodry EH, Karpati G, et al. Clinical phenotypes of different MPZ (P0) mutations may include Charcot-Marie-Tooth type IB, Déjérine-Sottas, and congenital hypomyelination. Neuron. 1996. 17:451–460.

73. Marrosu MG, Vaccargiu S, Marrosu G, Vannelli A, Cianchetti C, Muntoni F. Charcot-Marie-Tooth disease type 2 associated with mutation of the myelin protein zero gene. Neurology. 1998. 50:1397–1401.

74. Shy ME, Jani A, Krajewski K, Grandis M, Lewis RA, Li J, et al. Phenotypic clustering in MPZ mutations. Brain. 2004. 127:371–384.

75. Hattori N, Yamamoto M, Yoshihara T, Koike H, Nakagawa M, Yoshikawa H, et al. Demyelinating and axonal features of Charcot-Marie-Tooth disease with mutations of myelinrelated proteins (PMP22, MPZ and Cx32): a clinicopathological study of 205 Japanese patients. Brain. 2003. 126:134–151.

76. Lemke G, Axel R. Isolation and sequence of a cDNA encoding the major structural protein of peripheral myelin. Cell. 1985. 40:501–508.

77. Xu W, Manichella D, Jiang H, Vallat Jm, Lilien J, Baron P, et al. Absence of P0 leads to the dysregulation of myelin gene expression and myelin morphogenesis. J Neurosci Res. 2000. 60:714–724.

78. Wong MH, Filbin MT. The cytoplasmic domain of the myelin P0 protein influences the adhesive interactions of its extracellular domain. J Cell Biol. 1994. 126:1089–1097.

79. Carter J, Gragerov A, Konvicka K, Elder G, Weinstein H, Lazzarini RA. Neurofilament (NF) assembly; divergent characteristics of human and rodent NF-L subunits. J Biol Chem. 1998. 273:5101–5108.

80. Fabrizi GM, Cavallaro T, Angiari C, Bertolasi L, Cabrini I, Ferrarini M, et al. Giant axon and neurofilament accumulation in Charcot-Marie-Tooth disease type 2E. Neurology. 2004. 62:1429–1431.

81. De Jonghe P, Mersivanova I, Nelis E, Del Favero J, Martin JJ, van Broeckhoven C, et al. Further evidence that neurofilament light chain gene mutations can cause Charcot-Marie-Tooth disease type 2E. Ann Neurol. 2001. 49:245–249.

82. Zhu Q, Couillard-Despres S, Julien JP. Delayed maturation of regenerating myelinated axons in mice lacking neurofilaments. Exp Neurol. 1997. 148:299–316.

83. Kriz J, Zhu Q, Julien JP, Padjen AL. Electrophysiological properties of axons in mice lacking neurofilament subunit genes: disparity between conduction velocity and axon diameter in absence of NF-H. Brain Res. 2000. 885:32–44.

84. Jordanova A, De Jonghe P, Boerkoel CF, Takashima H, De Vriendt E, Ceuterick C, et al. Mutations in the neurofilament light chain gene (NEFL) cause early onset severe Charcot-Marie-Tooth disease type 1A. Brain. 2003. 126:590–597.

85. Choi BO, Lee MS, Shin SH, Hwang JH, Choi KG, Kim WK, et al. Mutational analysis of PMP22, MPZ, GJB1, EGR2 and NEFL in Korean Charcot-Marie-Tooth neuropathy patients. Hum Mutat. 2004. 24:185–186.
86. Herringham WP. Muscular atrophy of the peroneal type affecting many members of a family. Brain. 1889. 11:230–236.

87. Bergoffen J, Scherer SS, Wang S, Scott MO, Bone LJ, Paul DL, et al. Connexin mutations in X-linked Charcot-Marie-Tooth disease. Science. 1993. 262:2039–2042.

88. Birouk N, LeGuern E, Maisonobe T, Rouger H, Gouider R, Tardieu S, et al. X-linked Charcot-Marie-Tooth disease with connexin 32 mutations: clinical and electrophysiologic study. Neurology. 1998. 50:1074–1082.

89. Hahn AF, Bolton CF, White CM, Brown WF, Tuuha SE, Tan CC, et al. Genotype/phenotype correlations in Xlinked dominant Charcot-Marie-Tooth disease. Ann N Y Acad Sci. 1999. 883:366–382.

90. Fischbeck KH, ar-Rushdi N, Pericak-Vance M, Rozear M, Roses AD, Fryns JP. X-linked neuropathy: gene localization with DNA probes. Ann Neurol. 1986. 20:527–532.

91. Vital A, Ferrer X, Lagueny A, Vandenberghe A, Latour P, Goizet C, et al. Histopathological features of X-linked Charcot-Marie-Tooth disease in 8 patients from 6 families with different connexin32 mutations. J Peripher Nerv Syst. 2001. 6:79–84.

92. Nicholson SM, Gomes D, de Nechaud B, Bruzzone R. Altered gene expression in Schwann cells of connexin32 knockout animals. J Neurosci Res. 2001. 66:23–36.

93. Evans WH, Martin PE. Gap junctions: structure and function. Mol Membr Biol. 2002. 19:121–136.
94. Kumar NM, Gilula NB. Cloning and characterization of human and rat liver cDNAs coding for a gap junction protein. J Cell Biol. 1986. 103:767–776.

95. Scherer SS, Deschenes SM, Xu YT, Grinspan JB, Fischbeck KH, Paul DL. Connexin32 is a myelin-related protein in the PNS and CNS. J Neurosci. 1995. 15:8281–8294.

96. Rozear MP, Pericak-Vance MA, Fischbeck K, Stajich JM, Gaskell PC Jr, Krendel DA, et al. Hereditary motor and sensory neuropathy, X-linked: a half century follow-up. Neurology. 1987. 37:1460–1465.

97. Hahn AF, Brown WF, Koopman WJ, Feasby TE. X-linked dominant hereditary motor and sensory neuropathy. Brain. 1990. 113:1511–1525.

98. Bort S, Nelis E, Timmerman V, Sevilla T, Cruz-Martinez A, Martinez F, et al. Mutational analysis of the MPZ, PMP22 and Cx32 genes in patients of Spanish ancestry with Charcot-Marie-Tooth disease and hereditary neuropathy with liability to pressure palsies. Hum Genet. 1997. 99:746–754.

99. Silander K, Meretoja P, Juvonen V, Ignatius J, Pihko H, Saarinen A, et al. Spectrum of mutations in Finnish patients with Charcot-Marie-Tooth disease and related neuropathies. Hum Mutat. 1998. 12:59–68.

100. Mersiyanova IV, Ismailov SM, Polyakov AV, Dadali EL, Fedotov VP, Nelis E, et al. Screening for mutations in the peripheral myelin genes PMP22, MPZ and Cx32 (GJB1) in Russian Charcot-Marie-Tooth neuropathy patients. Hum Mutat. 2000. 15:340–347.

101. Mostacciuolo ML, Righetti E, Zortea M, Bosello V, Schiavon F, Vallo L, et al. Charcot-Marie-Tooth disease type 1 and related demyelinating neuropathies: mutation analysis in a large cohort of Italian families. Hum Mutat. 2001. 18:32–41.

102. Huehne K, Benes V, Thiel C, Kraus C, Kress W, Hoeltzenbein M, et al. Novel mutations in the Charcot-Marie-Tooth disease genes PMP22, MPZ, and GJB1. Hum Mutat. 2003. 21:100.

103. Yoshihara T, Yamamoto M, Doyu M, Mis KI, Hattori N, Hasegawa Y, et al. Mutations in the peripheral myelin protein zero and connexin32 genes detected by nonisotopic RNase cleavage assay and their phenotypes in Japanese patients with Charcot-Marie-Tooth disease. Hum Mutat. 2000. 16:177–178.

104. Numakura C, Lin C, Ikegami T, Guldberg P, Hayasaka K. Molecular analysis in Japanese patients with Charcot-Marie-Tooth disease: DGGE analysis for PMP22, MPZ, and Cx32/GJB1 mutations. Hum Mutat. 2002. 20:392–398.

105. Nelis E, Erdem S, Van Den Bergh PY, Belpaire-Dethiou MC, Ceuterick C, Van Gerwen V, et al. Mutations in GDAP1: autosomal recessive CMT with demyelination and axonopathy. Neurology. 2002. 59:1865–1872.

106. Baxter RV, Ben-Othmane K, Rochelle JM, Stajich JE, Hulette C, Dew-Knight S, et al. Ganglioside-induced differentiation-associated protein-1 is mutant in Charcot-Marie-Tooth disease type 4A/8q21. Nat Genet. 2002. 30:21–22.

107. Cuesta A, Pedrola L, Sevilla T, Garcia-Planells J, Chumillas MJ, Mayordomo F, et al. The gene encoding ganglioside-induced differentiation-associated protein 1 is mutated in axonal Charcot-Marie-Tooth type 4A disease. Nat Genet. 2002. 30:22–25.

108. Asbury AK, Gale MK, Cox SC, Baringer JR, Berg BO. Giant axonal neuropathy - a unique case with segmental neurofilamentous masses. Acta Neuropathol (Berl). 1972. 20:237–247.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download