Abstract
Background and Purpose
Encephalitis caused by Listeria monocytogenes (L. monocytogenes) is rare but sometimes fatal. Early diagnosis is difficult using routine cerebrospinal fluid (CSF) tests, while next-generation sequencing (NGS) is increasingly being used for the detection and characterization of pathogens.
Methods
This study set up and applied unbiased NGS to detect L. monocytogenes in CSF collected from three cases of clinically suspected listeria meningoencephalitis.
Results
Three cases of patients with acute/subacute meningoencephalitis are reported. Magnetic resonance imaging and blood cultures led to a suspected diagnosis of L. monocytogenes, while the CSF cultures were negative. Unbiased NGS of CSF identified and sequenced reads corresponding to L. monocytogenes in all three cases.
Conclusions
This is the first report highlighting the feasibility of applying NGS of CSF as a diagnostic method for central nervous system (CNS) L. monocytogenes infection. Routine application of this technology in clinical microbiology will significantly improve diagnostic methods for CNS infectious diseases.
Listeria monocytogenes (L. monocytogenes) is a Gram-positive bacterium with central nervous system (CNS) tropism that is an uncommon but important cause of severe CNS infections. L. monocytogenes infections can lead to meningitis, supratentorial abscesses (commonly in immunocompromised individuals) and even brainstem encephalitis (rhomboencephalitis) in immunocompetent patients in rare cases.12 Due to the high mortality and common serious sequelae in survivors, an accurate initial diagnostic approach is important for applying appropriate treatment early. Unfortunately, the early diagnosis of listeria encephalitis is a challenge to the clinician due to the deceptive findings for the cerebrospinal fluid (CSF) and the low positivity rate of the pathogen discovered by routine CSF Gram stains and cultures.234
Next-generation sequencing (NGS) is an emerging method with the potential to improve the ability to identify pathogens.5 However, there have been few reports on the use of NGS for the clinical diagnosis of CNS infectious diseases.6789 Here we present three clinically suspected cases of listeria meningoencephalitis with negative CSF cultures, which were finally confirmed by NGS of CSF.
Three patients with clinically suspected listeria meningoencephalitis admitted to Peking Union Medical College Hospital (PUMCH) from January 2014 to October 2014 were included in this study. Before administering antibiotics treatment, CSF was collected in accordance with standard procedures, snap frozen and stored at -20℃.
This study was approved by the Institutional Review Board of PUMCH and Beijing Genomics Institute, Shenzhen (IRB No. JS-890). The use of patients' clinical data and CSF samples for this study was approved by The Ethics Committee of PUMCH. Written informed consents were obtained from all patients or their legal surrogates.
DNA was extracted directly from the clinical samples using the TIANamp Micro DNA Kit (DP316, Tiangen Biotech, Beijing, China). DNA libraries were constructed through end-repaired adapter added overnight, and by applying polymerase chain reaction amplification to the extracted DNA. Quality control was carried out using a bioanalyzer (Agilent 2100, Agilent Technologies, Santa Clara, CA, USA) combined with PCR to measure the adapters before sequencing. DNA sequencing was then performed using the BGISEQ-100 platform (BGI-Tianjin, Tianjin, China).10
High-quality sequencing data were generated after filtering out low-quality, low-complexity, and shorter reads. To eliminate the effect of the human sequences, the data mapped to the human reference genome (hg19) were excluded using a powerful alignment tool called Burrows-Wheeler Alignment.11 The remaining data were then aligned to the Microbial Genome Database, which includes bacteria, viruses, fungi, and protozoa. Finally, the mapped data were processed by removing duplicate reads for advanced data analysis.
A nonredundant database that included all the published genomes of microorganisms was downloaded from the National Center for Biotechnology Information (ftp://ftp.ncbi.nlm.nih.gov/genomes/). The database used for this study contained 1,492 bacteria, 2,686 viruses, and 60 species of fungi that can cause infections in humans, and 33 species of protozoa related to human diseases. The depth and coverage of each species were calculated using SoapCoverage from the SOAP website (http://soap.genomics.org.cn/).
To validate the results of NGS, sequence-specific PCR identification of L. monocytogenes with a target fragment was carried out using the primers 5'-TATGTCGGGCAAGCG TTC-3' and 5'-GCGCTTGCGTGGTAATTC-3'. Sanger sequencing was performed with ABI PRISM 3730 DNA Sequencer (Applied Biosystems, Foster City, CA, USA) to validate the sequencing results. Finally, the sequences were aligned to the NT database with the online software NCBI Blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome).
The case series included three male adult patients aged from 40 to 66 years. All of them—who were previously otherwise healthy except for hypertension in Case 1—presented with an acute or a subacute onset. Two cases resulted in rhombencephalitis while one resulted in meningoencephalitis. The clinical characteristics of the three cases are summarized in Table 1. The most common prodromal symptoms included vertigo, nausea, vomiting, fever, and headache. Two of the cases experienced difficulty breathing and required mechanical ventilation, and the third progressed rapidly to coma. Cranial magnetic resonance imaging was performed in Cases 1 and 2 (Fig. 1). Both cases showed multiple irregular patchy and nodular lesions with T2 prolongation predominantly involving the brainstem and cerebellar pedunculus with patchy or ring enhancement. Lumbar punctures revealed increased opening pressure of the CSF in Cases 1 and 3, with a maximum of 260 mm H2O. Elevated cell counts and protein levels but normal CSF glucose levels were observed in all cases. A encephalitis virus antibody panel was negative for herpes simplex virus, cytomegalovirus, rubella virus, and toxoplasma gondii. CSF stains for bacteria, fungi, and acid-fast bacilli were negative. L. monocytogenes was cultured from blood but not from CSF in both Cases 1 and 2. As for Case 3, Gram-positive bacteria were found in a blood culture without being further identified, while the culture of his CSF was also negative (Table 2). All of the symptoms improved gradually after administering empirical treatment with intravenous antibiotics covering L. monocytogenes.
There were 19.1 million, 15 million, and 31 million total reads generated for Cases 1, 2, and 3, respectively; these quantities fulfill the analytical standard. After filtering out low-complexity and shorter reads, the reads were mapped to the human genome, which yielded 16.7 million, 13.5 million, and 28.6 million mapped reads for Cases 1, 2, and 3, respectively, with percentages of 93.92%, 95.35%, and 95.74% (Supplementary Table 1 in the online-only Data Supplement). The unmapped data that accounted for 5% of the cleaned data on average were then aligned to the Microbial Genome Database. The reads aligned to bacteria were different for each patient, with the percentage ranging from 0.05% to 0.28% (Supplementary Table 1 in the online-only Data Supplement). The number of reads aligned to viruses, fungi, and protozoa are also listed in Supplementary Table 1 (in the online-only Data Supplement).
The proportion of reads aligned to bacteria was higher than those mapped to viruses, fungi, and protozoa in almost all samples. We speculated that the three patients were suffering from bacterial infection. The coverage and depth of each species were calculated, and the species were sorted according to the coverage and the number of unique reads from high to low. Some species were excluded on the basis of their range of reference values, and some others (Supplementary Table 2 in the online-only Data Supplement) with high values were also excluded due to them being confirmed as contamination in our preliminary research (unpublished).
Finally, DNA of L. monocytogenes was identified in the CSF of the three patients. The identified number of unique reads mapped on the L. monocytogenes genome sequence ranged from 74 to 665, with percentages of 0.75–66.39%. The coverage of the identified L. monocytogenes gene on their genome varied from 0.28% to 2.4%, with depth values of 1.0 and 1.3, respectively. The number and percentage of unique reads, coverage, and depth on the identified L. monocytogenes DNA sequences are presented in Table 3 and Fig. 2.
Sequence-specific PCR identification for L. monocytogenes revealed the presence of L. monocytogenes in two samples (Supplementary Fig. 1 in the online-only Data Supplement). No sample for validation was available for Case 1.
Three clinically suspected cases of listeria meningoencephalitis with negative CSF cultures but confirmed by NGS were found in the present study. To our knowledge, this is the first study focusing on the application of NGS of CSF to the identification of L. monocytogenes.
Listeria encephalitis is a rare disease with a high mortality rate that is observed predominantly in previously healthy adults without a history of immunosuppression,2 as also found in the present study. Listeria encephalitis usually presents with a biphasic course. The prodromal symptoms include fever, headache, nausea, vomiting, and fever, and are then followed by the development of progressive neurological signs. Respiratory failure is commonly observed in cases of listeria rhomboencephalitis,2 as occurred in two of the present cases. The survival rate is greater than 70% if appropriate antibiotic therapy is started early.2 Prompt diagnosis and appropriate early antibiotic treatment are thus critical to an optimal outcome. However, the early diagnosis of this disease is challenging due to the deceptive CSF findings in these patients. First, the CSF findings in listeria rhombencephalitis often appears similar with that of viral meningoencephalitis.21213 The CSF profile is less likely to show an excessive white blood cell count or protein concentration as observed in our patient (Case 1). Second, L. monocytogenes is one of the few CNS pathogens associated with red blood cells in CSF, which cannot exclude the possibility of herpes simplex virus-1 encephalitis. Third, the Gram stains of CSF are negative in most cases, with a positivity rate of only 14%, due to its pleomorphism and its tendency to become over-decolorized.34 Finally, CSF and blood cultures have been reported to be positive only in 41% and 61% of cases, respectively.24 All of these features make an early diagnosis challenging and lead to a delay in appropriate treatment.
The sensitivity of conventional CSF tests of CNS infectious diseases has been low due to the diversity of pathogens and the absence of standardization among assays. An etiological diagnosis based on cultures, serological identification, and pathogen-specific PCR assays could be difficult because numerous pathogens can cause encephalitis. NGS has the potential to revolutionize the ability to identify common, rare, or even newly identified pathogens. Wilson et al.7 used unbiased NGS to identify Leptospira santarosai in the CSF of a case with meningoencephalitis in 2014, since when there has been great interest in the application of NGS technology for the comprehensive identification of infectious agents from CSF samples. Theoretically, according to the length of a long read, the number of hits to the microbial genome, the specific nucleic acid sequence and the completeness of reference database, almost all pathogens can be identified. The present study detected a significant amount of unique reads of the L. monocytogenes genome in the CSF of three patients. Moreover, no trace of these unique reads of L. monocytogenes was detected in control samples from three patients with autoimmune encephalitis processed concomitantly. Unique reads of Propionibacterium acnes and Micrococcus luteus were present in the control samples, which were confirmed in our preliminary research as environmental contamination from non-pathogenic DNA. The DNA sequence of L. monocytogenes identified by NGS was further confirmed by the PCR and Sanger methods in Cases 2 and 3. In addition, the diagnosis of L. monocytogenes infection made by NGS was consistent with the clinical symptoms and the CSF and neuroimaging evaluations. The favorable response to antibiotic therapy for L. monocytogenes further supported our diagnosis.
NGS has been found to be a rapid and precise method for the molecular diagnosis of simple to complex diseases. More than 43,000 microbial genomes have been released, comprising 38,000 bacteria and 5,000 viruses.14 These data also contain representatives of important human pathogens, and its availability will have great implications for the detection of microbial pathogens with the application of NGS.141516
This study highlights the practicability of deploying NGS of CSF as a rapid diagnostic assay for CNS L. monocytogenes infection. It can be predicted that its routine application to the panmicrobial identification of CSF could have a major impact on diagnosis approaches to CNS infectious diseases in the near future.
Figures and Tables
Fig. 1
Cranial magnetic resonance imaging of Case 1 (A and B) and Case 2 (C and D). A and B: Axial T2-weighted images showing irregular patchy hyperintense areas predominantly in the right lateral medullary and pons, and also in the cerebellar pedunculus. C: Sagittal T2-weighted image showing multiple patchy and nodular lesions with T2 prolongation involving the brainstem and upper cervical spinal cord. D: T1-weighted postcontrast image showing multiple parenchymal nodular and ring-enhanced lesions in the brainstem and upper cervical spinal cord.
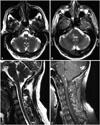
Fig. 2
NGS results of pathogen identification. A: In Case 1, 0.75% of bacterial reads corresponded to L. monocytogenes, with a coverage of 0.28%. B: In Case 2, 39.14% of bacterial reads corresponded to L. monocytogenes, with a coverage of 2.4%. C: In Case 3, 66.39% of bacterial reads corresponded to L. monocytogenes, with a coverage of 1.9%. L. monocytogenes: Listeria monocytogenes, NGS: next-generation sequencing.

Table 1
Clinical presentations of the three cases

Table 2
Routine laboratory evaluations of the three cases

Table 3
Number, percentage, coverage, and depth of unique reads for the sequences of Listeria monocytogenes in the CSF samples

| Case no. | Pathogen identified | No. of unique reads | Percentage, % | Coverage, % | Depth |
|---|---|---|---|---|---|
| 1 | L. monocytogenes | 74 | 0.75 | 0.28 | 1.0 |
| 2 | L. monocytogenes | 665 | 39.14 | 2.40 | 1.1 |
| 3 | L. monocytogenes | 486 | 66.39 | 1.90 | 1.3 |
Acknowledgements
We thank the physicians who provided clinical support. We also thank our patients and their families.
References
1. Workman S, Theal M. Rhomboencephalitis caused by Listeria monocytogenes. Can J Infect Dis. 1997; 8:113–116.
2. Armstrong RW, Fung PC. Brainstem encephalitis (rhombencephalitis) due to Listeria monocytogenes: case report and review. Clin Infect Dis. 1993; 16:689–702.

3. Mylonakis E, Hohmann EL, Calderwood SB. Central nervous system infection with Listeria monocytogenes. 33 years' experience at a general hospital and review of 776 episodes from the literature. Medicine (Baltimore). 1998; 77:313–336.

4. Paul ML, Dwyer DE, Chow C, Robson J, Chambers I, Eagles G, et al. Listeriosis--a review of eighty-four cases. Med J Aust. 1994; 160:489–493.
5. Long SW, Williams D, Valson C, Cantu CC, Cernoch P, Musser JM, et al. A genomic day in the life of a clinical microbiology laboratory. J Clin Microbiol. 2013; 51:1272–1277.

6. Grard G, Fair JN, Lee D, Slikas E, Steffen I, Muyembe JJ, et al. A novel rhabdovirus associated with acute hemorrhagic fever in central Africa. PLoS Pathog. 2012; 8:e1002924.

7. Wilson MR, Naccache SN, Samayoa E, Biagtan M, Bashir H, Yu G, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014; 370:2408–2417.

8. Briese T, Paweska JT, McMullan LK, Hutchison SK, Street C, Palacios G, et al. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 2009; 5:e1000455.

9. Yu G, Greninger AL, Isa P, Phan TG, Martínez MA, de la Luz Sanchez M, et al. Discovery of a novel polyomavirus in acute diarrheal samples from children. PLoS One. 2012; 7:e49449.

10. Jeon YJ, Zhou Y, Li Y, Guo Q, Chen J, Quan S, et al. The feasibility study of non-invasive fetal trisomy 18 and 21 detection with semiconductor sequencing platform. PLoS One. 2014; 9:e110240.

11. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760.

12. Reynaud L, Graf M, Gentile I, Cerini R, Ciampi R, Noce S, et al. A rare case of brainstem encephalitis by Listeria monocytogenes with isolated mesencephalic localization. Case report and review. Diagn Microbiol Infect Dis. 2007; 58:121–123.

13. Cunha BA, Fatehpuria R, Eisenstein LE. Listeria monocytogenes encephalitis mimicking Herpes Simplex virus encephalitis: the differential diagnostic importance of cerebrospinal fluid lactic acid levels. Heart Lung. 2007; 36:226–231.

14. Fournier PE, Dubourg G, Raoult D. Clinical detection and characterization of bacterial pathogens in the genomics era. Genome Med. 2014; 6:114.

Supplementary Materials
The online-only Data Supplement is available with this article at http://dx.doi.org/10.3988/jcn.2016.12.4.446.
Supplementary Table 2
The most common contaminated species in NGS results of CSF in our preliminary research
Supplementary Fig. 1
PCR result and Sanger sequencing result of pathogen validation. A: The Listeria monocytogenes sequence-specific PCR identification of case No. 2. The marker we used is Trans 2 K Plus DNA Maker. "M" and "N" represent marker and negative control, respectively. B: The Listeria monocytogenes sequence-specific PCR identification of case No. 3. The marker we used is DL2000 DNA Marker. "M" and "N" represent marker and negative control, respectively. C: The Sanger sequence of target fragments for case No. 2. D: The Sanger sequence of target fragments for case No. 3. PCR: polymerase chain reaction.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download