Abstract
Yeasts of the genus Malassezia are part of the normal flora of human skin. However, they are also associated with various skin diseases. Since the introduction of Malassezia to the Korean Dermatologic Society two decades ago, remarkable progress has been made in our knowledge of this genus. In this paper, we review recent developments in Malassezia research, including taxonomy and methods for species identification, recent genome analyses, Malassezia species distribution in healthy conditions and in specific skin diseases, trials investigating the mechanisms underlying Malassezia-related diseases, as well as therapeutic options. This review will enhance our understanding of Malassezia yeasts and related skin diseases in Korea.
Malassezia yeasts are lipophilic fungi that are part of the normal flora of the human skin. They are typically recovered from 75% to 98% of healthy adults12. However, these yeasts are also known to be associated with various diseases, including pityriasis versicolor (PV), seborrheic dermatitis, Malassezia folliculitis, and more recently, atopic dermatitis, which demonstrates their powerful allergenicity3.
Malassezia belongs to the kingdom Fungi, phylum Deuteromycota, class Blastomycetes, order Cryptococcales, and the family Cryptococcaceae4. The type species of the genus, Malassezia furfur (Robin) Babillon, was first described in 18894. Early taxonomic descriptions of Malassezia yeasts were limited by failure to culture these fungi, and thus, they were based solely on micromorphological descriptions in skin samples. Microbiological culture only became possible after the lipophilic nature of this fungus was uncovered5. Historically, Malassezia species were thought of as two separate organisms, a yeast (Pityrosporum) and a mycelial (Malassezia) fungus6. This dimorphism was finally resolved by Dorn and Roehnert7 and Porro et al.8, and since 1986, M. furfur is the name that has been formally accepted for both growth phases9. Using the medium validated by Leeming and Notman10, Cunningham et al.11 classified M. furfur isolates into three different cultural groups, which correspond to the three serological groups A, B, and C. Prior to 1996, the genus Malassezia comprised three taxa, M. furfur (serotype A, B, C), M. pachydermatis, and M. sympodialis 111213. In 1996, Guého et al.12 isolated four new species, M. globosa, M. obtusa, M. restricta, and M. slooffiae on the basis of morphology, ultrastructure, physiology, and molecular biology (Fig. 1, 2, 3). Between 2002 and 2004, four new species, M. dermatis14, M. japonica15, M. yamatoensis16, and M. nana17, were identified by Japanese scientists using molecular analyses, and since then, three more species, M. caprae18, M. equina18, and M. cuniculi19, have been identified in animals (Fig. 4). Thus, the genus Malassezia currently comprises 14 species, and new species continue to be discovered.
In Korea, various attempts have been made to identify members of this species. In 1996, Ahn and Ashbee20 introduced Malassezia to the Korean Dermatologic Society and compared the antifungal activities of azoles against M. furfur serovars A, B, and C by determining their minimal inhibitory concentrations using Leeming and Notman medium.
Malassezia yeasts have been implicated in various skin diseases, including PV, seborrheic dermatitis, Malassezia folliculitis, atopic dermatitis, and psoriasis (Fig. 5)2. An association between Malassezia yeasts and skin diseases was first reported by Eichstedt21 in 1846, and was based on the presence of yeasts and filaments in material from the infected scales of patients with PV. In Korea, correlations between various skin diseases and Malassezia species have been reported since 1997. After Guého et al.12 classified Malassezia into seven species in 1996, Ahn22 cultured the yeast from PV lesions to comply with the newly revised taxonomy, and reported that M. globosa was the most common causative agent of PV in Korea. Studies on the incidence of Malassezia species in acneiform eruptions23, steroid acne23, atopic dermatitis24, and seborrheic dermatitis24 were reported in 1998.
In 1999, Kang and Kim25 investigated the frequency of Malassezia yeasts in the comedones of patients clinically diagnosed with acne vulgaris and found that 25% of these patients had Malassezia folliculitis; however, no species-specific relationship could be identified. Kim et al.26 performed a skin prick test and measured total immunoglobulin E (IgE) and specific IgE antibodies to M. furfur in patients with head and neck dermatitis (HND). Of the 80 patients with HND, 45% were positive for M. furfur in the skin prick test, and 68% had specific IgE antibodies to M. furfur. The patients who showed a positive response to anti-M. furfur-specific IgE antibodies had more severe clinical symptoms and had higher total IgE levels. In conjunction with other environmental factors, such as sweat, heat, dryness, sun exposure, and stress, M. furfur was shown to aggravate HND.
Colonization of neonates and infants with Malassezia yeasts is a controversial topic. Oh et al.27 found that 60.5% (121/200) of neonates and infants under 12 weeks carried Malassezia yeasts in at least one of five examined sites: scalp, forehead, cheek, earwax, and back. Ahn et al.28 confirmed the efficacy of a one-week regimen of itraconazole by studying 20 patients with PV, and found that M. globosa infection was closely related to PV.
With the aim of obtaining baseline data, several studies have investigated the distribution of Malassezia species on normal human skin according to body region using swab and scrub-wash techniques293031. The incidence of Malassezia yeasts was 78.4% on the scalp, 86.5% on the forehead, 100% on the chest, and 97.3% on the back31. In normal subjects, M. restricta was found predominantly on the forehead and scalp, whereas M. globosa was predominantly present on the chest and back31.
In 2001, Lee et al.32 found that M. restricta was also the most frequently recovered species from the face of patients with seborrheic dermatitis. A 2002 study aimed at determining the clinical features of acne associated with Malassezia as well as the efficacy of antifungal treatments, showed that Malassezia-associated acne was characterized by polymorphous eruptions composed of open and closed comedones, inflammatory papules, and pustules, accompanied by seborrhea and seborrheic dermatitis. Malassezia-associated acne was also found to be aggravated during the summer, because of the use of systemic corticosteroids, and during menstruation. A one-week regimen of systemic itraconazole (200 mg/d) improved the lesions33.
In 2003, Lee et al.34 found that the efficacy and safety of 1.5% ciclopirox olamine (a hydroxypyridone derivative) shampoo was comparable to that of 2% ketoconazole shampoo (a highly effective antifungal treatment) in patients with mild-to-moderate dandruff. Moreover, in 2003, Jang et al.35 found that M. restricta and M. globosa are the most common causative species for Malassezia folliculitis of the face and trunk, respectively.
Subsequently, in 2004, Choe et al.36 performed a study on the distribution by age group and body site of Malassezia yeasts on normal human skin. Subjects between 21 and 30 years old showed the highest positive culture rate (88%), and the chest (91%) and thigh (60%) had respectively the highest and lowest positive culture rates of all evaluated body parts. M. globosa was most frequently recovered from the chest, whereas M. restricta was most frequently recovered from the forehead. Jang et al.37 conducted a study on earwax in 2005, and showed that M. restricta is the most commonly isolated species.
Most of the aforementioned studies were qualitative. However, in 2006, Lee et al.38 conducted a quantitative study on the distribution of Malassezia species in a healthy Korean population. In this study, the qualitative distribution of Malassezia species varied according to anatomic site: M. restricta was most commonly found on the scalp and forehead, whereas M. globosa was the predominant species on the chest. Quantitative analysis showed that, although the values varied among age groups, the yeast count per unit area of skin (cm2) was higher for the chest and scalp and lower for the upper arm and thigh. By age, the Malassezia yeast count on the scalp, forehead, and chest was the highest in the 11 to 20 and 21 to 30 years age groups.
In 2009, Moon et al.39 compared Malassezia species from atopic dermatitis head and neck lesions in children and adults and showed that M. globosa was the predominant species in children, whereas M. furfur was the predominant species in adults. Both Pityrosporum ovale-specific IgE and clinical severity grade were higher in the adults, and they were significantly correlated.
In 2014, Song et al.40 performed a retrospective study of patients with folliculitis who were previously diagnosed with Malassezia folliculitis or non-Malassezia folliculitis and compared their clinical features. Among the 80 cases of non-Malassezia folliculitis, yeasts were retrospectively found in 16 patients by serial tissue sectioning and diastase periodic acid Schiff staining. Therefore, the diagnosis of these cases was changed to Malassezia folliculitis. Malassezia folliculitis presented with trunk involvement and male predilection.
In order to overcome the limitations of conventional approaches based on morphology, microstructure analysis, and physiology for the delineation of closely related Malassezia species, molecular methods have been developed for the reliable isolation of Malassezia41424344454647484950. For the last 15 years, the molecular studies on Malassezia species in Asia have been performed mainly in Japan141516174647484950. Japanese scientists have designed non-culture-based methods, including the direct application of OpSite transparent dressings47. These methods avoid culture in Leeming and Notman agarose gels, as DNA is extracted directly from the dressings, followed by gene sequencing and nested polymerase chain reaction (PCR) for species identification and strain typing. The preferred targets for strain identification are the D1/D2 region of the 26S ribosomal DNA (rDNA), the internal transcribed spacer (ITS) regions, and the intergenic spacer 1 region of the ribosomal RNA gene (Fig. 6A). Quantitative analyses of Malassezia species have also been performed using real-time (RT) PCR4950.
In Korea, Lee et al.51 were the first to use 26S rDNA PCR and restriction fragment length polymorphism (PCR-RFLP) to identify Malassezia species in 2006 (Fig. 6B, 7). In 2007, new experimental molecular methods were introduced in Korea to identify and classify Malassezia species. Lim et al.52 successfully isolated M. dermatis from healthy subjects and patients with seborrheic dermatitis by using 26S rDNA PCR-RFLP and 26S rDNA and ITS1 sequencing for the first time in Korea (Fig. 8). Kim et al.53 implemented a colony PCR method that eliminated the need for DNA extraction, as a fast and simple way to amplify Malassezia target DNA, and assessed its clinical utility. In this method, Malassezia colonies were lysed in a microwave and used as a template for PCR instead of purified genomic DNA, and this colony PCR method was compared to two DNA extraction methods (the boiling and glass bead methods). Song et al.54 employed pyrosequencing to identify Malassezia yeasts, but the result showed limitations (Fig. 9). Lim et al.55 performed a nested PCR to differentiate various Malassezia species obtained from clinical strains and skin scales in 2008 (Fig. 10, 11). The detection rate for the nested PCR method was 96% for clinical strains and 87% for skin scales. M. globosa, M. sympodialis, and M. restricta were the most common causative agents in patients with PV, Malassezia folliculitis, and seborrheic dermatitis, respectively. Lee et al.56 isolated 19 strains of M. dermatis from healthy human skin in Korea by using 26S rDNA PCR-RFLP.
Using the same technique, in 2009, Jang et al.57 attempted the qualitative isolation of Malassezia yeasts from 80 healthy males and 80 healthy females in different age groups, from different body areas. They isolated M. dermatis and found that M. restricta, M. globosa, and M. sympodialis were the most commonly identified species. In addition, in 2009, Oh et al.58 compared PCR-RFLP and nested PCR for the isolation of Malassezia species and showed that PCR-RFLP is the preferred method for differentiation, although nested PCR is advantageous with respect to time and simplicity (Fig. 12). In 2009, there was also a special trial to study the effects of detergents on the morphology and immunomodulatory activity of M. furfur. In this study, Kim et al.59 found that detergent altered the surface of M. furfur and that detergent-treated M. furfur induced higher levels of TNF-α expression in monocytes than untreated M. furfur. They concluded that the detergents in shampoos or soaps affect the lipid layers of the Malassezia cell walls, which can induce or aggravate some inflammatory conditions.
The disorders associated with Malassezia yeasts have also been studied extensively in Korea. In 2010, Yim et al.60 demonstrated evidence of a relationship between M. sympodialis and atopic dermatitis by showing that it is the dominant species in patients with this disorder. However, in 2010, Oh et al.61 showed that there was no statistically significant difference in the distribution of Malassezia species between patients with seborrheic dermatitis and healthy controls. Kim et al.62 developed and evaluated the accuracy of a multiplex PCR kit using ITS1 specific primers with six Malassezia standard strains.
In 2011, Song et al.63 detected Malassezia yeasts in acne patients using 26S rDNA PCR-RFLP. The authors showed that the growth rate of Malassezia was clearly lower in patients with acne (50%) than in controls (70.6%) and that M. restricta was the dominant species in patients with acne, whereas M. globosa was the most common species found in healthy controls. Using the same method, Lee et al.64 investigated the distribution of Malassezia species on the scalp of patients with seborrheic dermatitis. The most common species in these patients was M. restricta, whereas M. globosa was most common in healthy controls, which suggests that M. restricta is the most important Malassezia species in Korean patients with seborrheic dermatitis.
In 2012, Lim et al.65 compared the yields of Malassezia DNA isolated by the conventional culture-based technique or by a non-culture-based technique via OpSite adhesive tape developed by Sugita et al.47. They found that the culture rate of the non-culture-based technique was better than that of the culture-based technique (100% vs. 57.8%); the only disadvantage of the non-culture-based technique was the need for repeated rounds of PCR because of the low amounts of extracted DNA.
A recent genome analysis of M. globosa revealed that the absence of a gene encoding fatty acid synthase might be compensated by genes encoding lipases and phospholipases, which showed higher expression levels on the human scalp66. Based on this finding, in 2013, Lee et al.67 analyzed the in vivo expression of the lipases and phospholipases of M. restricta, the most frequently isolated Malassezia species from the scalp of patients with seborrheic dermatitis in Korea, by two-step nested RT-PCR. Another study in 2013 examined the use of pyrosequencing for the identification of Malassezia species68. Using different primers targeting ITS1 and ITS2, designed in a previous study by Song et al.54, Malassezia species were successfully isolated by pyrosequencing, and this method was more rapid and accurate than 26S rDNA PCR-RFLP.
In 2011, Lee et al.69 measured the changes in transepidermal water loss (TEWL), stratum corneum hydration, and skin pH in PV lesions of 11 patients. They found significantly higher TEWL and reduced hydration in lesional skin in comparison to in the adjacent non-lesional skin; there was no change in skin pH. The authors concluded that infection with Malassezia species alters the biophysical properties of skin, especially the function of the stratum corneum as a barrier to water loss.
Park et al.70 investigated the skin characteristics of patients with PV using MPA5® (Courage and Khazaka, Köln, Germany). Both hyperpigmented and hypopigmented PV lesions showed higher humidity, sebum levels, and TEWL than healthy controls, indicating that higher humidity and sebum levels provide a better environment for Malassezia yeasts in the skin, leading to disruption of the skin barrier, which causes a further increase in TEWL.
Regarding alternative therapies for Malassezia-associated skin diseases, Lee et al.71 conducted a pilot study on the efficacy of methyl 5-amino-levulinic acid photodynamic therapy for recalcitrant Malassezia folliculitis, and showed that it is an effective treatment option. In 2012, Wi et al.72 examined the antifungal effect of light-emitting diode (LED) irradiation on M. furfur, M. sympodialis, and M. globosa. They found that an LED that emitted light at wavelengths of 380±2 nm and 392.5±1 nm had an antifungal effect on Malassezia species, and they observed an increase in both intracellular and extracellular reactive oxygen species following LED irradiation at 392.5±1 nm. In addition, Kim et al.73 reported the efficacy of a shampoo with a new formula that contains natural ingredients, including an extract of Rosa centifolia petals and epigallocatechin gallate, which is known to exert anti-inflammatory and antifungal effects on scalp seborrheic dermatitis. In a randomized double-blind controlled study, the shampoo had an efficacy comparable to that of 1% zinc pyrithione shampoo and of 2% ketoconazole shampoo; thus, it could be used as an alternative treatment for seborrheic dermatitis.
Many common skin diseases, such as seborrheic dermatitis, atopic dermatitis, and Malassezia folliculitis, are associated with Malassezia yeasts. Although these yeasts are part of the normal flora of human skin, under certain conditions, they can induce or aggravate skin diseases. Many studies have tried to elucidate the mechanism and role of Malassezia in disease by comparing the distribution of Malassezia in specific diseases to that in healthy controls. These efforts have been supported by the development of highly accurate molecular methods for the identification of each species. In addition, several studies have described the clinical manifestation of related diseases and have investigated various therapeutic options. Recent genomic analyses of Malassezia species have accelerated the elucidation of the mechanisms underlying these skin diseases. More prolific studies are underway in Korea, in sync with the global trend.
Figures and Tables
 | Fig. 1 |
 | Fig. 2Malassezia globosa. (A) Medium-sized, lighter in color, friable and crenated flat colonies with a pointed button center (Leeming and Notman medium, 34℃, 14 days). (B) Spherical, circular cells with buds on a narrow base (Parker Quink-KOH stain, ×1,000). Data from the article of Ahn (Korean J Med Mycol 1998;3:81-88)2. |
 | Fig. 3Malassezia restrica. (A) Small-sized, circular, umbonate, entire, dull colonies (Leeming and Notman medium, 34℃, 14 days). (B) Small, spherical or oval cells with buds on a relatively narrow base (Parker Quink-KOH stain, ×1,000). Data from the article of Ahn (Korean J Med Mycol 1998;3:81-88)2. |
 | Fig. 4Malassezia dermatis. (A) Large-sized, circular, smooth colonies (Leeming and Notman medium, 34℃, 14 days). (B) Spherical, oval, or ellipsoidal vegetative cells with monopolar budding (Parker Quink-KOH stain, ×1,000). Data from the article of Lim et al. (Korean J Dermatol 2007;45:1020-1030)52. |
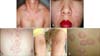 | Fig. 5Skin diseases associated with Malassezia species. (A) Pityriasis versicolor, (B) seborrheic dermatitis, (C) Malassezia folliculitis, (D) atopic dermatitis, and (E) psoriasis. |
 | Fig. 6(A) Schematic representation of the rRNA gene in the type strain (CBS 7966) of Malassezia globosa: the 26S rDNA, internal transcribed spacer (ITS1) region, and intergenic spacer 1 (IGS1) region sequences are used for species identification and strain typing. Data from the article of Sugita et al. (J Clin Microbiol 2003;41:3022-3027)48. (B) Sequences of the amplified 26S rDNA products from clinical isolates of Malassezia. Data from the article of Lee et al. (Korean J Med Mycol 2006;11:141-153)51. |
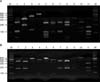 | Fig. 7Polymerase chain reaction (PCR) and restriction fragment length polymorphism patterns of the 26S rDNA PCR products digested with (A) Hha1 and (B) BstF51 from Malassezia standard strains. Lane M, 100-bp DNA ladder; Lane 1, Malassezia furfur; Lane 2, M. sympodialis; Lane 3, M. globosa; Lane 4, M. restricta; Lane 5, M. slooffiae; Lane 6, M. pachydermatis; Lane 7, M. japonica; Lane 8, M. nana; Lane 9, M. dermatis; Lane 10, M. obtusa; and Lane 11, M. yamatoensis. Data from the article of Lee et al. (Korean J Med Mycol 2006;11:141-153)51. |
 | Fig. 8A molecular phylogenetic tree constructed by the neighbor-joining method using the internal transcribed spacer 1 sequences of members of the genus Malassezia. Data from the article of Lim et al. (Korean J Dermatol 2007;45:1020-1030)52. |
 | Fig. 9Sequence alignment of the rRNA internal transcribed spacer 2 variable regions (box) of 11 Malassezia standard strains for pyrosequencing. Lane 1, Malassezia dermatis; Lane 2, M. furfur; Lane 3, M. globosa; Lane 4, M. japonica; Lane 5, M. nana; Lane 6, M. obtusa; Lane 7, M. pachydermatis; Lane 8, M. restricta; Lane 9, M. slooffiae; Lane 10, M. sympodialis; and Lane 11, M. yamatoensis. Data from the article of Song et al. (Korean J Med Mycol 2007;12:189-197)54. |
 | Fig. 10Nested polymerase chain reaction: structure of the internal transcribed spacer (ITS) gene region and location of primer sites. Data from the articles of Lim et al. (Korean J Dermatol 2008;46:446-452)55. |
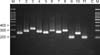 | Fig. 11Nested polymerase chain reaction products from standard Malassezia species. Lane M, molecular marker; Lane 1, Malassezia dermatis; Lane 2, M. furfur; Lane 3, M. globosa; Lane 4, M. japonica; Lane 5, M. nana; 6, M. obtusa; Lane 7, M. pachydermatis; Lane 8, M. restricta; Lane 9, M. slooffiae; Lane 10, M. sympodialis; Lane 11, M. yamatoensis; and Lane C, negative control. Data from the article of Lim et al. (Korean J Dermatol 2008;46:446-452)55. |
 | Fig. 12Flowchart of the nested polymerase chain reaction (PCR) and restriction fragment length polymorphism (PCR-RFLP) methods. Data from the article of Oh et al. (Ann Dermatol 2009;21:352-357)58. |
References
1. Kundu RV, Garg Amit. Yeast infections: Candidiasis, Tinea (Pityriasis) Versicolor, and Malassezia (Pityrosporum) Folliculitis. In : Goldsmith LA, Katz SI, Gilchrest BA, Paller AS, Leffell DJ, Wolff K, editors. Fitzpatrick's dermatology in general medicine. 8th ed. New York: McGraw-Hill;2012. p. 2298–2311.
2. Ahn KJ. Taxonomy of the genus Malassezia. Korean J Med Mycol. 1998; 3:81–88.
3. Gupta AK, Batra R, Bluhm R, Boekhout T, Dawson TL Jr. Skin diseases associated with Malassezia species. J Am Acad Dermatol. 2004; 51:785–798.

4. Kwon-Chung KJ, Bennett JE. Medical mycology. . Philadelphia: Lea & Febiger;1992. p. 170–182.
5. Benham RW. The cultural characteristics of Pityrosporum ovale-a lipophilic fungus. J Invest Dermatol. 1939; 2:187–203.
7. Dorn M, Roehnert K. Dimorphism of Pityrosporum orbiculare in a defined culture medium. J Invest Dermatol. 1977; 69:244–248.
8. Porro MN, Passi S, Caprilli F, Mercantini R. Induction of hyphae in cultures of Pityrosporum by cholesterol and cholesterol esters. J Invest Dermatol. 1977; 69:531–534.

9. Cannon PF. International Commission on the Taxonomy of Fungi (ICTF): name changes in fungi of microbiological, industrial and medical importance. Part 2. Microbiol Sci. 1986; 3:285–287.
10. Leeming JP, Notman FH. Improved methods for isolation and enumeration of Malassezia furfur from human skin. J Clin Microbiol. 1987; 25:2017–2019.

11. Cunningham AC, Leeming JP, Ingham E, Gowland G. Differentiation of three serovars of Malassezia furfur. J Appl Bacteriol. 1990; 68:439–446.
12. Guého E, Midgley G, Guillot J. The genus Malassezia with description of four new species. Antonie Van Leeuwenhoek. 1996; 69:337–355.

14. Sugita T, Takashima M, Shinoda T, Suto H, Unno T, Tsuboi R, et al. New yeast species, Malassezia dermatis, isolated from patients with atopic dermatitis. J Clin Microbiol. 2002; 40:1363–1367.

15. Sugita T, Takashima M, Kodama M, Tsuboi R, Nishikawa A. Description of a new yeast species, Malassezia japonica, and its detection in patients with atopic dermatitis and healthy subjects. J Clin Microbiol. 2003; 41:4695–4699.

16. Sugita T, Tajima M, Takashima M, Amaya M, Saito M, Tsuboi R, et al. A new yeast, Malassezia yamatoensis, isolated from a patient with seborrheic dermatitis, and its distribution in patients and healthy subjects. Microbiol Immunol. 2004; 48:579–583.

17. Hirai A, Kano R, Makimura K, Duarte ER, Hamdan JS, Lachance MA, et al. Malassezia nana sp. nov., a novel lipid-dependent yeast species isolated from animals. Int J Syst Evol Microbiol. 2004; 54:623–627.

18. Cabañes FJ, Theelen B, Castellá G, Boekhout T. Two new lipid-dependent Malassezia species from domestic animals. FEMS Yeast Res. 2007; 7:1064–1076.
19. Cabañes FJ, Vega S, Castellá G. Malassezia cuniculi sp. nov., a novel yeast species isolated from rabbit skin. Med Mycol. 2011; 49:40–48.

20. Ahn KJ, Ashbee HR. Determination of minimum inhibitory concentrations of several azole antifungals for Malassezia furfur. Ann Dermatol. 1996; 8:187–194.

21. Eichstedt E. Pilzbildung in der Pityriasis versicolor. Frorip Neue Notizen aus dem Gebeite der Naturkunde Heilkinde. 1846; 39:270–271.
22. Ahn KJ. Malassezia species cultured from the lesions of pityriasis versicolor. Korean J Dermatol. 1997; 35:736–743.
23. Yu HJ, Kim YS, Yang HY, Kim JH, Lee SK, Son SJ. The incidences of Malassezia in steroid acne and other acneiform eruptions. Korean J Med Mycol. 1998; 3:24–32.
24. Hwang SJ, Lee SH, Kim DW, Jun JB, Chung SL, Jung HJ. Comparison of clinical manifestations and distribution of Malassezia species in patients with seborrheic dermatitis and atopic dermatitis. Korean J Dermatol. 1998; 36:617–627.
25. Kang SH, Kim HU. The isolation of Malassezia yeasts in the comedones of acne vulgaris. Korean J Med Mycol. 1999; 4:33–39.
26. Kim TY, Jang IG, Park YM, Kim HO, Kim CW. Head and neck dermatitis: the role of Malassezia furfur, topical steroid use and environmental factors in its causation. Clin Exp Dermatol. 1999; 24:226–231.

27. Oh CK, Kwon KS, Lee CW, Cho SH, Jang HS, Park JH. The distribution of Malassezia yeasts on the skin of neonates and infants. Korean J Med Mycol. 1999; 4:27–32.
28. Ahn KJ, Kim KJ, Yi GJ. Efficacy of one-week regimen of itraconazole for pityriasis versicolor. Korean J Med Mycol. 1999; 4:124–130.
29. Kwon HC, Kang SH, Kim HU. The distribution of Malassezia yeasts on normal human skin by culture study using the scrub-wash techique. Korean J Dermatol. 1999; 37:38–45.
30. Kwon HC, Kang SH, Kim HU. The distibution of Malassezia yeasts on normal human skin by culture study using the swabbing technique. Korean J Dermatol. 1999; 37:46–56.
31. Kim SC, Kim HU. The distribution of Malassezia species on the normal human skin according to body region. Korean J Med Mycol. 2000; 5:120–128.
32. Lee YW, Kang HJ, Ahn KJ. Malassezia species cultured from the lesions of seborrheic dermatitis. Korean J Med Mycol. 2001; 6:70–76.
33. Youn NH, Cha SH, Park SD. Malassezia yeasts in acne vulgaris. Korean J Dermatol. 2002; 40:1453–1460.
34. Lee JH, Lee HS, Eun HC, Cho KH. Successful treatment of dandruff with 1.5% ciclopirox olamine shampoo in Korea. J Dermatolog Treat. 2003; 14:212–215.

35. Jang SJ, Choi YB, Ahn KJ. Malassezia species cultured from the lesions of Malassezia folliculitis. Korean J Med Mycol. 2003; 8:55–62.
36. Choe YB, Jang SJ, Yim SM, Ahn KJ. The quantitative study on the distribution of Malassezia yeasts on the normal skin of the young adults. Korean J Med Mycol. 2004; 9:174–181.
37. Jang SJ, Yim SM, Lee YW, Choe YB, Ahn KJ. The distribution of Malassezia yeasts on the earwax of young adults. Korean J Dermatol. 2005; 43:1583–1589.
38. Lee YW, Yim SM, Lim SH, Choe YB, Ahn KJ. Quantitative investigation on the distribution of Malassezia species on healthy human skin in Korea. Mycoses. 2006; 49:405–410.

39. Moon HS, Son SJ, Park K, Chae JD. Difference of Malassezia species and Pityrosporum ovale specific IgE in head and neck lesions of atopic dermatitis related to ages and severity. Korean J Med Mycol. 2009; 14:1–8.
40. Song HS, Kim SK, Kim YC. Comparison between Malassezia folliculitis and non-Malassezia folliculitis. Ann Dermatol. 2014; 26:598–602.
41. Senczek D, Siesenop U, Böhm KH. Characterization of Malassezia species by means of phenotypic characteristics and detection of electrophoretic karyotypes by pulsed-field gel electrophoresis (PFGE). Mycoses. 1999; 42:409–414.
42. Guillot J, Deville M, Berthelemy M, Provost F, Guého E. A single PCR-restriction endonuclease analysis for rapid identification of Malassezia species. Lett Appl Microbiol. 2000; 31:400–403.

43. Theelen B, Silvestri M, Guého E, van Belkum A, Boekhout T. Identification and typing of Malassezia yeasts using amplified fragment length polymorphism (AFLP), random amplified polymorphic DNA (RAPD) and denaturing gradient gel electrophoresis (DGGE). FEMS Yeast Res. 2001; 1:79–86.

44. Gaitanis G, Velegraki A, Frangoulis E, Mitroussia A, Tsigonia A, Tzimogianni A, et al. Identification of Malassezia species from patient skin scales by PCR-RFLP. Clin Microbiol Infect. 2002; 8:162–173.

45. Gupta AK, Kohli Y, Summerbell RC. Molecular differentiation of seven Malassezia species. J Clin Microbiol. 2000; 38:1869–1875.

46. Makimura K, Tamura Y, Kudo M, Uchida K, Saito H, Yamaguchi H. Species identification and strain typing of Malassezia species stock strains and clinical isolates based on the DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J Med Microbiol. 2000; 49:29–35.

47. Sugita T, Suto H, Unno T, Tsuboi R, Ogawa H, Shinoda T, et al. Molecular analysis of Malassezia microflora on the skin of atopic dermatitis patients and healthy subjects. J Clin Microbiol. 2001; 39:3486–3490.

48. Sugita T, Kodama M, Saito M, Ito T, Kato Y, Tsuboi R, et al. Sequence diversity of the intergenic spacer region of the rRNA gene of Malassezia globosa colonizing the skin of patients with atopic dermatitis and healthy individuals. J Clin Microbiol. 2003; 41:3022–3027.

49. Sugita T, Tajima M, Tsubuku H, Tsuboi R, Nishikawa A. Quantitative analysis of cutaneous Malassezia in atopic dermatitis patients using real-time PCR. Microbiol Immunol. 2006; 50:549–552.

50. Tajima M, Sugita T, Nishikawa A, Tsuboi R. Molecular analysis of Malassezia microflora in seborrheic dermatitis patients: comparison with other diseases and healthy subjects. J Invest Dermatol. 2008; 128:345–351.

51. Lee YW, Lim SH, Ahn KJ. The application of 26S rDNA PCR-RFLP in the identification and classification of Malassezia yeast. Korean J Med Mycol. 2006; 11:141–153.
52. Lim SH, Kim SM, Jung BR, Lee YW, Choe YB, Ahn KJ. A mycological and molecular biological study of Malassezia dermatis isolated from Korean. Korean J Dermatol. 2007; 45:1020–1030.
53. Kim SM, Lim SH, Jung BR, Lee YW, Choe YB, Ahn KJ. The application of colony PCR in the molecular biological analysis of Malassezia yeasts. Korean J Med Mycol. 2007; 12:180–188.
54. Song YC, Lim SH, Jung BR, Lee YW, Choe YB, Ahn KJ. The application of pyrosequencing method in the identification and classification of Malassezia yeasts. Korean J Med Mycol. 2007; 12:189–197.
55. Lim SW, Shin MG, Lim JY, Yun SJ, Kim SJ, Lee SC, et al. Nested PCR for detection of Malassezia species from patient skin scales and clinical strains. Korean J Dermatol. 2008; 46:446–452.
56. Lee YW, Kim SM, Oh BH, Lim SH, Choe YB, Ahn KJ. Isolation of 19 strains of Malassezia dermatis from healthy human skin in Korea. J Dermatol. 2008; 35:772–777.

57. Jang SJ, Lim SH, Ko JH, Oh BH, Kim SM, Song YC, et al. The investigation on the distribution of Malassezia yeasts on the normal Korean skin by 26S rDNA PCR-RFLP. Ann Dermatol. 2009; 21:18–26.

58. Oh BH, Song YC, Lee YW, Choe YB, Ahn KJ. Comparison of nested PCR and RFLP for identification and classification of Malassezia yeasts from Healthy human skin. Ann Dermatol. 2009; 21:352–357.

59. Kim SH, Ko HC, Kim MB, Kwon KS, Oh CK. The effect of detergents on the morphology and immunomodulatory activity of Malassezia furfur. Ann Dermatol. 2009; 21:130–135.

60. Yim SM, Kim JY, Ko JH, Lee YW, Choe YB, Ahn KJ. Molecular analysis of Malassezia microflora on the skin of the patients with atopic dermatitis. Ann Dermatol. 2010; 22:41–47.

61. Oh BH, Lee YW, Choe YB, Ahn KJ. Epidemiologic study of Malassezia yeasts in seborrheic dermatitis patients by the analysis of 26S rDNA PCR-RFLP. Ann Dermatol. 2010; 22:149–155.

62. Kim HM, Lim YY, Park EJ, Chun YJ, Kim MN, Kim BJ, et al. The development and evaluation of multiplex PCR technique for identification of Malassezia Yeast. Korean J Med Mycol. 2010; 15:51–60.
63. Song YC, Hahn HJ, Kim JY, Ko JH, Lee YW, Choe YB, et al. Epidemiologic study of Malassezia yeasts in acne patients by analysis of 26S rDNA PCR-RFLP. Ann Dermatol. 2011; 23:321–328.

64. Lee YW, Byun HJ, Kim BJ, Kim DH, Lim YY, Lee JW, et al. Distribution of Malassezia species on the scalp in Korean seborrheic dermatitis patients. Ann Dermatol. 2011; 23:156–161.

65. Lim SH, Kim YR, Jung JW, Hahn HJ, Lee YW, Choe YB, et al. A comparison study between culture based technique and Op-site non-culture based technique for identifying Malassezia yeasts on normal skin. Korean J Med Mycol. 2012; 17:217–229.

66. DeAngelis YM, Saunders CW, Johnstone KR, Reeder NL, Coleman CG, Kaczvinsky JR Jr, et al. Isolation and expression of a Malassezia globosa lipase gene, LIP1. J Invest Dermatol. 2007; 127:2138–2146.
67. Lee YW, Lee SY, Lee Y, Jung WH. Evaluation of expression of lipases and phospholipases of Malassezia restricta in patients with seborrheic dermatitis. Ann Dermatol. 2013; 25:310–314.

68. Kim JY, Hahn HJ, Choe YB, Lee YW, Ahn KJ, Moon KC. Molecular biological identification of Malassezia yeasts using pyrosequencing. Ann Dermatol. 2013; 25:73–79.

69. Lee WJ, Kim JY, Song CH, Jung HD, Lee SH, Lee SJ, et al. Disruption of barrier function in dermatophytosis and pityriasis versicolor. J Dermatol. 2011; 38:1049–1053.

70. Park HJ, Lee YW, Choe YB, Ahn KJ. Skin characteristics in patients with pityriasis versicolor using non-invasive method, MPA5. Ann Dermatol. 2012; 24:444–452.

71. Lee JW, Kim BJ, Kim MN. Photodynamic therapy: new treatment for recalcitrant Malassezia folliculitis. Lasers Surg Med. 2010; 42:192–196.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download