Abstract
Background
Recently, identification of fungi have been supplemented by molecular tools, such as ribosomal internal transcribed spacer (ITS) sequence analysis. According to these tools, morphological Exophiala species was newly introduced or redefined.
Objective
This study was designed to investigate the phylogenetics based on ribosomal ITS sequence analysis from clinical Exophiala species isolated in Korea.
Methods
The strains of Exophiala species were 4 clinical isolates of phaeohyphomycosis agents kept in the department of dermatology, Dongguk University Medical Center(DUMC), Gyeongju, Korea. The DNAs of total 5 strains of Exophiala species were extracted by bead-beating method. Polymerase chain reaction of ITS region using the primer pairs ITS1-ITS4, was done and phylogenetic tree contributed from sequences of ITS region from 5 Korean isolates including E. dermatitidis CBS 109154 and comparative related strains deposited in GenBank.
Results
The strains of Exophiala species were 3 strains of E. dermatitidis, 1 strain of E. jeanselmei and 1 strain of Exophiala new species. Among the 3 subtypes (type A, B, C) of E. jeanselmei, E. jeanselmei DUMC 9901 belonged to type B. Of the 2 main types of E. dermatitidis (type A, B) and 3 subtypes of E. dermatitidis type A (A0, A1 and A2), two strains (E. dermatitidis CBS 709.95, E. dermatitidis CBS 109154) belonged to A0 subtypes, 1 strain (E. dermatitidis DUMC 9902) A1 subtype, respectively.
Exophiala is the main genus of black yeasts, characterised by annellidic conidiogenesis and mostly isolated from environmental substrates, including soil, wood, and other plant material. The majority of these infections are cutaneous and subcutaneous, but fatal systemic infections can occur1,2. Genus Exophiala includes E. jeanselmei complex, E. dermatitidis and E. spinifera complex. E. jeanselmei complex has darkened rocket-shaped conidiogenous cells without multicellular conidiophores. E. spinifera, unlike E. jeanselmei, has large multicellular conidiophores and capsular material around budding cells. E. dermatitidis has numerous conidiophores and conidiogenous cells either intercalary or free, and flask shaped. This species grows at up to 42℃, shows no growth with nitrate and nitrite and is sometimes called Wangiella dermatitidis1. With recent advances in molecular biological techniques such as internal transcribed spacer (ITS) sequences analysis and phylogenetic analysis, Exophiala species has been further classified and new species have been identified and named3-15. As a result, E. jeanselmei have recently been molecular biologically re-identified as including E. jeanselmei, E. xenobiotica, E. oligosperma, E. exophialae, E. nishimurae, E. bergeri and E. nigra5. E. jeanselmei have been further classified as fifteen subtypes by ITS-restriction fragment length polymorephism (RFLP) analysis8. E. dermatitidis also have been subgrouped as A, B, C or D. Most of the subgroup A contains clinical strains, whereas subgroup B contains environmental strains11. For this reason ITS sequences analysis and phylogenetic analysis have remained a very useful distinguishing parameter for Exophiala species.
This study was designed to determine the molecular phylogenetics of Exophilala species isolated from phaeohyphomycosis patients in Korea.
In this study five clinical strains, which had been isolated from the phaeohyphomycosis patients, were included: two strains of E. jeanselmei (E. jeanselmei Dongguk University Medical Center [DUMC] 9901 and E. jeanselmei DUMC 0501) and two strains of E. dermatitidis (E. dermatitidis DUMC 9902 and E. dermatitidis CBS 709.95), both isolated from four phaeohyphomycosis patients and preserved in our hospital, and one strain of E. dermatitidis CBS 109154 isolated from a patient with a cerebral infection which was obtained from GenBank (Table 1). The identification and typing of the Korean isolates were confirmed by colony morphology, microscopy, sugar assimilation test, heat tolerance test, and the ITS sequence analysis.
Fungi grown at 25℃ in Sabouraud's dextrose agar for 2 weeks were mixed with glass beads (0.5 mm diameter) in distilled water and shaken for 5 minutes. To purify DNA, the mixture was extracted with phenol/chloroform/isoamyl alcohol (25:24:1) (Sigma, St. Louis, MO, USA) and centrifuged at 12,000 rpm at room temperature for 10 minutes. A tenth volume of 3 M sodium acetate (Sigma) and 3 volumes of absolute ethanol were added to the supernatant for DNA precipitation at -20℃ for 12 hours. DNA was then centrifuged, rinsed with 70% ethanol, dried, and stored at -20℃ in distilled water.
To amplify the ITS 1-5.8S rDNA-ITS 2 region of rDNA according to White et al.16, universal primers ITS1 (5'-TCCGTAGGTGAACCTGCGG-3') and ITS4 (5'-TCCTC CGCTTATTGATATGC-3') were prepared by Bioneer Corp. (Daejeon, Korea). Fungal DNA was mixed with 10× PCR buffer, 1.6 ml of 2.5 mM dNTP, 0.4 ml of primers and 5 units of Taq polymerase (Takara, Otsu, Japan). After heating at 95℃ for 3 minutes, 30 cycles of denaturation at 95℃ for 30 seconds, annealing at 60℃ for 30 seconds, and extension at 72℃ for 1 minute were performed and then followed by the final extension at 72℃ for 10 minutes using a thermal cycler, PTC-100 (MJ Research Inc., Watertown, MA, USA). The amplified DNA was electrophoresed on a 1% agarose gel containing ethidium bromide at 100 volt for 20~30 minutes using a Mupid-2 Mini Gel System (Cosmo Bio Co., Seoul, Korea). The amplified DNA fragments were observed under ultraviolet light, cut out of the gel, purified using AccuPrep® PCR Purification Kit (Bioneer Corp.), and then subjected to DNA sequencing by Macrogen Inc. (Seoul, Korea).
After the ITS sequences of the four strains of Exophiala species isolated from Korea were determined, GenBank was searched using the Blast program to find identical or similar sequences. Thereafter, the ITS sequences of E. dermatitidis CBS 109154 and relative strains that represent each subgroup of Exophiala species were obtained from GenBank for the alignment of ITS sequences by using Clustal X version 2.017. The ITS sequences were aligned by Neighbor-Jointing (NJ) analysis using MEGA4 software18 with Close-Neighbor-Interchange algorithm with an option of search level 7 and 1,000 booststrap replicates19. All alignment gaps that were introduced to maximize the homology were considered missing data, and the branch distances were calculated by using the average pathway method19.
The ITS regions of four Korean isolates of Exophiala species preserved in our hospital were amplified to produce approximately 643-bp fragments. After the ITS sequences of E. dermatitidis CBS 109154 and relative strains that represent each subgroup of Exophiala species were obtained, the length of ITS nucleotides was compared by multiple alignment of ITS sequences. The number of ITS nucleotides was around 552 in E. jeanselmei and around 584 in E. dermatitidis. There were almost no significant differences in length.
A phylogenetic tree was constructed by NJ analysis, and the evolutionary disturbances between individual strains were described as horizontal branches. The ITS sequences of twenty five Exophiala strains including the five Korean isolates and twenty representative strains were compared. The five Korean isolates did not show morphological diversity and only three species, including one strain of E. jeanselmei, three strains of E. dermatitidis and one strain of other Exophiala species were identified. E. jeanselmei DUMC O501 did not show any identical ITS sequences to Exophiala species, which was regarded as a new species (Fig. 1).
E. jeanselmei was classified into three subtypes: type A, which showed identical ITS sequneces to the type strain, E. jeanselmei CBS 507.90, and type B and C which were not. Kawasaki et al.8 have reported intraspecies variation of the genotypes of E. jeanselmei isolated from patients. To compare with subtypes of this study, we treated with suppositive restriction enzyme. It is presumed that types A and B identified by this study would be identical with E5 identified by Kawasaki et al.8 and that type C identified by this study would be identical with E2 or E3. E. jeanselmei DUMC 9901 belonged to the type B reported in Japan and United States, and showed identical ITS sequences with the Japanese species E. jeanselmei CBS 116.86. Korean strains and Japanese strains caused only skin infections, although there is a lack of information on human infections (Table 2, Fig. 2).
Matos et al.11 subclassified E. dermatitidis as group A (clinical strain), group B (environmental strain), group C or group D. In this study, we also subclassified the isolates as groups A, B and D. Since the ITS sequences of group C was not available, group C was excluded. Group A was further divided into 3 subgroups: group A0 which was identical with E. dermatitidis CBS 207.35, and groups A1 and A2 which were similar to E. dermatitidis CBS 207.35. All three Korean isolates were in group A. E. dermatitidis CBS 709.95 and E. dermatitidis CBS 10915 belonged to group A0, and E. dermatitidis DUMC 9902 belonged to A1. Since groups A0 and A1 have been isolated in many countries, including Japan, China, United States and Germany, there is no significant regional difference in isolated strains between countries. Human infections ranged in virulence from skin infections to fatal systemic infections (Table 3, Fig. 3).
Exophiala species invades the human body, mainly causing phaeohyphomycosis which shows brown hyphae or yeast-like spores in involved tissue. The species rarely causes either chromoblastomycosis in which round-shaped sclerotic cells or muriform cells with a thick wall are observed or eumycotic mycetoma which is characterized by the development of abscesses, draining sinuses, and the formation of fistulae discharging granules1,20,21. Phaeohyphomycosis usually occurs in the skin or subcutaneous tissue. The skin lesion appears as pustules or verrucous plaques. It is rarely disseminated to the internal organs22-24. In Korea, three strains of E. jeanselmei species and three strains of E. dermatitidis were isolated from patients with phaeohyphomycosis25-30. Most of these strains caused subcutaneous infections, but only one strain of E. dermatitidis species involved the brain30. More strains of Exophiala species were isolated in many countries: 188 strains in United States6, 76 strains in Japan9 and 20 strains in China3. It is expected that more strains will appear in Korea. In this study, we used two strains of E. jeanselmei and two strains of E. dermatitidis which were isolated and preserved in our hospital as well as one strain of E. dermatitidis CBS 109154 obtained from GenBank. One strain of E. jeanselmei isolated by Kim et al.27 was excluded from the study because no molecular biological information was available. Of these five strains, one was identified as E. jeanselmei by Suh et al.25 through morphological analysis. However, this strain was regarded as a new strain because it had no identical ITS sequences with Exophiala species. Therefore, molecular biological analysis is recommended as a supplementary method if morphological analysis is inadequate.
The classification and identification of Exophiala species have been performed by morphological examination, including colony morphology and light microscopy, as well as a physiological examination, including sugar assimilation tests and heat tolerance1. Molecular biological analysis has recently been used as a supplementary method3-15. E. jeanselmei species is morphologically and molecular biologically heterogeneous4-10,13,14, whereas E. dermatitidis species is homogeneous6,8,9,14.
In the past, E. jeanselmei species were re-identified as E. jeanselmei var. jeanselmei, E. jeanselmei var. heteromorpha and E. jeanselmei var. lecanii-corni by morphological and cultural features31. After then, this species were further classified as E. jeanselmei var. jeanselmei, E. jeanselmei var. heteromorpha and E. jeanselmei var. lecanii-corni and E. lecanii-corni by mitochondrial DNA analysis14. Furthermore, E. jeanselmei was newly subclassified E. jeanselmei as E. jeanselmei, E. xenobiotica, E. oligosperma, E. lecanii-corni and E. heteromorpha by sequence analysis of the ITS regions, elongation factor 1-α (EF1-α) and β-tubulin (β-TUB)7. Currently, E. jeanselmei is much more subdivided into seven species, including E. jeanselmei, E. xenobiotica, E. oligosperma, E. exophialae, E. nishimurae, E. bergeri and E. nigra by sequence analysis of the ITS regions5. The above-mentioned species was confirmed as fifteen subtypes by ITS-RFLP analysis8.
In this study, we classified E. jeanselmei as types A, B and C by ITS-RFLP analysis, and matched with fifteen subtypes suggested by Kawasaki et al.8; types A and B are considered to be subtype E5, whereas type C is considered to be subtype E2 or E3. In addition, since E. jeanselmei DUMC 9901 corresponds to type B, and subtype E5 is the most commonly isolated subtype in Japan, there is a possibility that more type B strains will be identified in Korea.
Although the DUMC O501 strain isolated in Korea was identified as E. jeanselmei by morphological and physiological analyses, it had a 84% (432/512) homology to E. jeanselmei by ITS sequences analysis. While this strain was close to Pseudocladosporium species and E. salmonis, this is regarded as a new strain because no identical ITS sequences were detected from GenBank. Since most of the strains with similar ITS sequences originate from soil, this strain is also thought to originate from soil. Because morphological and physiological analyses have some limitations in the identification of strains, molecular biological analysis is recommended as supplementary method. Since one of the five Korean isolates is a new species, further new species will be identified in Korea.
Since E. dermatitidis species is very homogeneous6,8,9,14, it had not been subclassified by morphological or molecular biological analysis before the studies by Uijthof et al.15 which reported that of the five subgroups, group I is most common and the number of nucleotides in groups II to V is different by 1 to 4 from that in group I. In 2003, Matos et al.11 further classified E. dermatitidis species as groups A, B, C and D by ITS sequence analysis and M-13 fingerprinting. They also stated that most strains belong to groups A and B, and that group A strains are clinical isolates, whereas group B strains are environmental isolates. In this study, it was found that the 3 strains isolated in Korea were classified as A0, A1 and A2, although there were no significant differences in locations and clinical features between A0, A1 and A2.
In conclusion, the five Korean isolates did not show more diversity than western isolates and only three species, including one strain of E. jeanselmei, three strains of E. dermatitidis and one strain of other Exophiala species were identified. All these strains were isolated from patients with phaeohyphomycosis. Despite the low number of the strains included in this study, E. dermatitidis is the most commonly isolated strains from phaeohyphomycosis patients in Korea. In the Western world, however, E. jeanselmei is the main causative agent of phaeohyphomycosis.
Taken together, ITS sequence analysis and phylogenetic analysis can supplement traditional morphological and physiological analyses in the identification of Exophiala species as well as the evaluation of its distribution, determination of subtypes and detection of new species.
Figures and Tables
Fig. 1
Neighbor-joining tree based on sequences of the ITS region from the 25 members of Exophiala species and relatives; neighbor-joining algorithm with 1,000 bootstrap replicates. R. aquaspersa CBS 313.73 is taken as an outgroup. ITS: internal transcribed spacer, E.: Exophiala, R.: Rhinocladiella, sp.: species, DUMC: Dongguk University Medical Center.

Fig. 2
Neighbor-joining tree based on sequences of the ITS region from the 11 members of E. jeanselmei and relatives; neighbor-joining algorithm with 1,000 bootstrap replicates. E. spinifera CBS 899.68T is taken as an outgroup. ITS: internal transcribed spacer, E.: Exophiala.
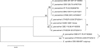
Fig. 3
Neighbor-joining tree based on sequences of the ITS region from the 15 members of E. dermatitidis and relatives; neighbor-joining algorithm with 1,000 bootstrap relatives E. spinifera CBS 899.68T is taken as an outgroup. ITS: internal transcribed spacer, E.: Exophiala.

Fig. 4
Neighbor-joining tree based on sequences of the ITS region from the 9 members of Exophiala species DUMC 0501 (new species) and relatives; neighbor-joining algorithm with 1,000 bootstrap replicates. ITS: internal transcribed spacer, DUMC: Dongguk University Medical Center, E.: Exophiala, u.: uncultured, f.: fungal.

References
1. De Hoog GS, Guarro J, Gene J, Figueras MJ. Atlas of clinical fungi. 2000. 2nd ed. Virgili: Centraalbureau voor Schimmelcultures;645–668.
2. Ellis D, Davis S, Alexiou H, Handke R, Bartley R. Descriptions of medical fungi. 2007. 2nd ed. Adelaide: Nexus Print Solutions;62–65.
3. Li DM, Li RY, De Hoog GS, Wang YX, Wang DL. Exophiala asiatica, a new species from a fatal case in China. Med Mycol. 2009. 47:101–109.

4. Aoyama Y, Nomura M, Yamanaka S, Ogawa Y, Kitajima Y. Subcutaneous phaeohyphomycosis caused by Exophiala xenobiotica in a non-Hodgkin lymphoma patient. Med Mycol. 2009. 47:95–99.

5. Zeng JS, De Hoog GS. Exophiala spinifera and its allies: diagnostics from morphology to DNA barcoding. Med Mycol. 2008. 46:193–208.

6. Zeng JS, Sutton DA, Fothergill AW, Rinaldi MG, Harrak MJ, de Hoog GS. Spectrum of clinically relevant Exophiala species in the United States. J Clin Microbiol. 2007. 45:3713–3720.

7. De Hoog GS, Zeng JS, Harrak MJ, Sutton DA. Exophiala xenobiotica sp. nov., an opportunistic black yeast inhabiting environments rich in hydrocarbons. Antonie Van Leeuwenhoek. 2006. 90:257–268.

8. Kawasaki M, Anzawa K, Tanabe H, Mochizuki T, Ishizaki H, Nishimura K. Intra-species variation of genotypes of Exophiala jeanselmei isolated from patients in Japan. Nihon Ishinkin Gakkai Zasshi. 2005. 46:261–265.

9. Kawasaki M, Ishizaki H, Nishimura K, Miyaji M. Mitochondrial DNA analysis of Exophiala jeanselmei and Exophiala dermatitidis. Mycopathologia. 1990. 110:107–112.

10. de Hoog GS, Vicente V, Caligiorne RB, Kantarcioglu S, Tintelnot K, Gerrits van den Ende AH, et al. Species diversity and polymorphism in the Exophiala spinifera clade containing opportunistic black yeast-like fungi. J Clin Microbiol. 2003. 41:4767–4778.

11. Matos T, Haase G, Gerrits van den Ende AH, de Hoog GS. Molecular diversity of oligotrophic and neurotropic members of the black yeast genus Exophiala, with accent on E. dermatitidis. Antonie Van Leeuwenhoek. 2003. 83:293–303.
12. Vitale RG, de Hoog GS. Molecular diversity, new species and antifungal susceptibilities in the Exophiala spinifera clade. Med Mycol. 2002. 40:545–556.

13. Wang L, Yokoyama K, Miyaji M, Nishimura K. Identification, classification, and phylogeny of the pathogenic species Exophiala jeanselmei and related species by mitochondrial cytochrome b gene analysis. J Clin Microbiol. 2001. 39:4462–4467.

14. Kawasaki M, Ishizaki H, Matsumoto T, Matsuda T, Nishimura K, Miyaji M. Mitochondrial DNA analysis of Exophiala jeanselmei var. lecanii-corni and Exophiala castellanii. Mycopathologia. 1999. 146:75–77.
15. Uijthof JM, Van Belkum A, De Hoog GS, Haase G. Exophiala dermatitidis and Sarcinomyces phaeomuriformis: ITS1-sequencing and nutritional physiology. Med Mycol. 1998. 36:143–151.

16. White TJ, Burns T, Lee S, Taylor J. Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. Amplication and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. 1990. San Diego: Academic Press;315–322.

17. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997. 25:4876–4882.

18. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007. 24:1596–1599.

19. Nei M, Kumar S. Molecular evolution and phylogenetics. 2000. New York: Oxford University Press.
20. Queiroz-Telles F, Esterre P, Perez-Blanco M, Vitale RG, Salgado CG, Bonifaz A. Chromoblastomycosis: an overview of clinical manifestations, diagnosis and treatment. Med Mycol. 2009. 47:3–15.

21. Al-Tawfiq JA, Amr SS. Madura leg due to Exophiala jeanselmei successfully treated with surgery and itraconazole therapy. Med Mycol. 2009. 47:648–652.

24. Suh MK, Lee YH. Infectious caused by dematiaceous fungi. Korean J Med Mycol. 2005. 10:77–82.
25. Suh MK, Kwon SW, Kim TH, Sun YW, Lim JW, Ha GY, et al. A case of subcutaneous phaeohyphomycosis caused by exophiala jeanselmei. Korean J Dermatol. 2005. 43:124–127.
26. Suh MK, Suh JC, Seo SK, Na GY, Kim YJ, Bang JS, et al. A case of subcutaneous phaeohyphomycosis caused by exophiala jeanselmei. Korean J Dermatol. 1999. 37:395–399.
27. Kim HU, Kang SH, Matsumoto T. Subcutaneous phaeohyphomycosis caused by Exophiala jeanselmei in a patient with advanced tuberculosis. Br J Dermatol. 1998. 138:351–353.

28. Kim DS, Yoon YM, Kim SW. Phaeohyphomycosis due to. Exophiala dermatitidis successfully treated with itraconazole. Korean J Med Mycol. 1999. 4:79–83.
29. Lee SC, Chun IK, Kim YP. A case of phaeomycotic subcutaneous abscess caused by Wangiella Dermatitidis. Korean J Dermatol. 1986. 24:692–696.
30. Chang CL, Kim DS, Park DJ, Kim HJ, Lee CH, Shin JH. Acute cerebral phaeohyphomycosis due to Wangiella dermatitidis accompanied by cerebrospinal fluid eosinophilia. J Clin Microbiol. 2000. 38:1965–1966.

31. De Hoog GS. Rhinocladiella and allied genera. Stud Mycol. 1977. 15:141–144.




 PDF
PDF ePub
ePub Citation
Citation Print
Print






 XML Download
XML Download