Abstract
Background
The expression of c-Met is substantially elevated in most malignant human cancers. We therefore searched for c-Met expression and compared the expression level among malignant skin cancers.
Objective
The aim of this study was to determine the c-Met expression pattern and the protein expression level in selected malignant cutaneous tumors.
Methods
G361 cells (malignant melanoma cell line) and A431 cells (squamous cell carcinoma cell line) were cultured and analyzed, using immunoprecipitation and Western blot analysis, for expression of c-Met. Additionally, 16 separate specimens of malignant melanomas (MMs), 16 squamous cell carcinomas (SCCs), 16 basal cell carcinomas (BCCs) and 16 normal tissues were analyzed for the expression of c-Met using immunohistochemical studies.
Results
Based on cultured cell immunoprecipitation and Western blot analysis, both A431 cells and G361 cells expressed c-Met, however, c-Met was expressed substantially more in G361 cells. Immunohistochemical examination of c-Met showed that it was over-expressed in all malignant skin cancers. In addition, c-Met expression was more increased in MM compared to other cancers.
Hepatocyte growth factor (HGF) is a well known growth factor that has been implicated to be involved in the mitogenic process, cell motility, angiogenesis and epithelial morphogenesis, while playing a pleiotropic role in other biological processes such as normal development, wound healing, and carcinogenesis1-5. c-Met receptor (170 kD) is a growth factor receptor, formed by two polypeptide chains of α-subunit (50 kD) and β-subunit (145 kD) linked by disulfide bonds. These chains make up an extracellular ligand-binding domain, a hydrophobic transmembrane segment and an intracellular tyrosine kinase domain5. Generally, it has been understood that c-Met is located in cytoplasmic portion of the cell. But, in recent studies, it was reported that c-Met can be located or translocated to the nucleus in some cancer cells6,7. C-Met was initially identified as the protein product of a transforming oncogene8,9. C-Met pathway is activated by HGF, which then increases cellular mobilization and invasiveness4,10. There are some reports that show over-expression of c-Met in a number of human cancers, including thyroid, pancreas, stomach, prostate, colon, ovary, breast, kidney, liver and endometrial cancers5-22. Some cancers, such as gastric cancers and thyroid cancers, have been shown to have a relationship between c-Met expression with tumor stage and poor prognosis12,13. One study reported that metastatic melanomas had an increased level of c-Met expression among melanocytic lineage lesions13. In this study, we examined c-Met location and c-Met expression in human malignant skin cancers.
Cancer specimens were obtained from patients who had undergone surgery between January 2000 and October 2009, in the Departments of Dermatology and Plastic and Reconstructive Surgery at the Soonchunhyang University Hospital. The normal skin tissues were collected from the backs of 16 women who had breast reconstruction with latissimus dorsi flap. For immunohistochemical studies, archival formalin-fixed, paraffin-embedded tissues were used. The specimens consisted of 16 samples of malignant melanomas (MMs), 16 squamous cell carcinomas (SCCs), 16 basal cell carcinomas (BCCs) and 16 normal human skin tissues.
The human malignant melanoma cell line G361 and human squamous cell carcinoma cell lines A431 were cultured in DMEM, 10% FCS, 100 U/ml penicillin, 100 mg/ml streptomycin at 37℃, 5% CO2.
Cytoplasmic and nuclear extracts were prepared according to the instructions of the NE-PER® nuclear and cytoplasmic extraction kit (Pierce, Rockford, IL, USA). The fractions were then stored at -80℃ and used for Western blot analysis.
For immunoprecipitation, 500µl of cell extract (~1.0 mg/ml protein) was incubated with 1µl of affinity purified rabbit polyclonal c-Met antibody (SC-161; Santa Cruz Biotechnology, Santa Cruz, CA, USA) 10 mg/ml, at 4℃ overnight, followed by the addition of 30µl of Protein A/G PLUS-Agarose (SC-2003; Santa Cruz Biotechnology) for another 3 hr with shaking. The immunoprecipitates were collected by centrifugation, washed three times with 0.5 M LiCl, once with 1 ml of solution B (20 mM Tris-HCI at pH 7.0, 0.5 mM dithiothreitol, 1 mM phenylmethanesulfonylfluoride 1 mM benzamidine, and 0.5 mg/ml aprotinin).
Proteins from the subcellular fraction and immunoprecipitates were separated on NuPAGE 4~12% bis-Tris polyacrylamide gels (Invitrogen, Camarillo, CA, USA) and then electrophoretically transferred to Immuno-Blot PVDF membranes. The membranes were then incubated for 1 hr at room temperature, with a 1:500 dilution of rabbit anti-c-Met antibody (Santa Cruz Biotechnology). Next, horseradish peroxidase-conjugated secondary antibody (Cell signaling Technologies, Beverly, MA, USA) was applied at a dilution of 1:5,000. Anti-β-actin antibody was used as a loading control and the signal was visualized using an ECL detection kit (Amersham Biosciences, Amersham, Buckinghamshire, UK).
For immunohistochemical analysis, Parraffin sections (4µm) were prepared and staining was performed by the ABC method. In brief, slides were deparaffinized in xylene and rehydrated with ethanol. The endogenous peroxidase activity was inhibited by immersion of the slides in 3% H2O2/methanol. Antigen retrieval was performed in a microwave oven for 15 min with 10 mM citrate buffer (pH 6.0). Pre-incubation took place with a blocking solution for 30 min, to avoid unspecific binding. The sections were then incubated over night at 4℃ with the primary c-Met-specific polyclonal antibody (SC-161; Santa Cruz Biotechnology, 1:100). The slides were consecutively incubated with biotinylated secondary antibody for 30 min and then for 30 min with streptavidin-peroxidase. The visualization of the immunoreaction was performed with 3,3'-Diaminobenzidine (DBC500; ScyTek, Logan, UT, USA). Negative controls were performed as previously described, substituting the primary antibody with phosphate buffered saline.
In immunohistochemical evaluation, the histochemical score (HSCORE) was used for comparison and standardization23-25. The HSCORE of c-Met was determined by two sets of independent investigators. In c-Met stain, cells with brown cytoplasm under an optical microscope meant positive. HSCORE was calculated using the following equation: HSCORE ΣPi(i+1), where i is the intensity of staining with a value of 0, 1 or 2 (0, weak staining; 1, moderate staining or 2, strong staining) and Pi is the percentage of stained tumor cells varying from 0% to 100% (0.0~1.00). HSCOREs ranged from a minimum of zero in cases with no staining to a maximum of 3.0 in cases in which all the tumor cells were stained with maximal intensity. The percentage of stained tumor cells were calculated by counting positively stained cells among 500 tumor cells, using 10×10 grid in 400 magnified view.
The results of the Western blot analysis showed that c-Met was expressed in both cell lines-G361 cells and A431 cells. The c-Met was located within the cytoplasmic fraction of both cells (Fig. 1).
The staining pattern of c-Met in MM, SCC and BCC could be observed (Fig. 2). Immunohistochemical study further supported the Western blot analysis that c-Met expression was increased in MM and SCC (Table 1). The c-Met was over-expressed in MMs. The HSCORE ranged from 1.60 to 2.70. The mean HSCORE was 2.29 (standard deviation (SD)=0.301), indicating a very strong expression of c-Met. The HSCORE for SCCs ranged from 0.80 to 1.70, with the average at 1.17 (SD=0.279). In BCCs, the c-Met was weakly expressed. The HSCORE ranged from 0.50 to 0.80 (mean 0.64, SD=0.078). Immunohistochemistry results showed that c-Met was strongly expressed in MMs, while it was moderately expressed in SCCs and slightly expressed in BCCs, thereby revealing statistically significant differences among these cancers (p<0.001). However, in normal human skin tissues, c-Met was not well-expressed (Fig. 3). MMs have stronger positive responses than do other cancers and normal skin tissues. Indeed, deeper invasive melanoma tissues are observed to have more increased positive responses to the immunohistochemicals among theses MM tissues (Fig. 4).
There are now over 75 known human receptor tyrosine kinases (RTK), and many of them are reported to induce alterations in signal transduction molecular pathways4,10,26. They are known as proto-oncogenes involved in oncogenesis and tumor progression mechanisms. Examples of such RTK proto-oncogenes are EGF, c-kit, PDGF, Flt3 and c-Met10,26. c-Met was originally identified as an activated oncogene protein involved in a chromosomal translocation in a human osteogenic sarcoma cell line treated with N-methyl-N-nitro-N-nitrosoguanidine11. Activation of HGF/c-Met signal pathways has been known to promote cell motility, morphogenesis, wound healing and tissue regeneration. However, c-Met is also expressed in a variety of malignant cells. In an animal model study, the over expression of wild-type Met among hepatocytes was identified to be sufficient enough to cause hepatocellular carcinomas and c-Met mutations also have been reported in various other cancers27,28. Germ line missense mutations of c-Met, found in hereditary papillary renal carcinomas, provides compelling evidence that c-Met has a direct role in human cancers. Met-activating mutations are also found in gastric cancer and hepatocellular carcinomas26,28. In addition to these cancers, there are many reports identifying increased c-Met expression in other cancers such as colon cancers, dermatofibroma sarcoma protuberans, breast cancers, prostate cancers, endometrial cancers, ovarian cancers, lung cancers and head and neck cancers.
Studies surrounding the expression of c-Met in skin cancers are rare. There is one paper which observes the expression of c-Met in melanomas and melanocytic lineages. In a study researching the expression of c-Met in human melanocytic lesions, c-Met receptor expression was detected not in benign melanocytic lesions but in melanomas, especially metastatic melanomas13. It has been suggested that c-Met expression may be correlated with metastatic progression. In a study observing the distribution of HGF in human tissues, HGF was observed not only in active mitochemical cell populations, but also in mitotically quiescent and non-dividing cells. Indeed, almost all growth factors are found at low concentrations in various cells, but high affinity binding between growth factors and its receptors are required to induce the specific signals29. This implies that c-Met, the HGF receptor, is important to the signal transduction process. It has been well-known that c-Met is located in the cytoplasm, however, interestingly, in recent studies it was found that c-Met can be translocated to the nucleus either by HGF stimulation or without HGF6. c-Met cytoplasmic fragments were present in the nuclei especially in MDA-MB231 breast carcinoma cells6. But, in other studies with SkHep1 cells, c-Met was found only in the cytoplasmic portion7. To identify c-Met in malignant skin cancer cells, we separated the cytoplasmic portion and nucleus portion of G361 cells and A431 cells; as a result, c-Met was found to be expressed in the cytoplasm just like in SkHep1 cells.
In order to identify the role of c-Met in skin cancers, we examined malignant skin cancers, including BCCs, SCCs and MMs. Our results showed that, based on immunohistochemical stains, all skin cancers display increased c-Met expression. MMs have a stronger positive response than other cancers. Protein expressions corresponded to our results. Immunohistochemically, deeper MMs were observed to express c-Met more strongly than superficial ones.
C-Met is thought to be related to the malignancy potential. MM is characterized as one of the most invasive cancers. In a prostate cancer study, mitogenic activity of HGF increased in a dose dependent manner and c-Met expression was increased according to the HGF, suggesting that a complex of autocrine, paracrine and endocrine trophic or mitogenic pathways may be involved in the progression of cancer invasion30. These results imply that c-Met may be implicated, in part, in tumor progression (invasion or metastasis) and that the difference of HGF receptors on tumor cells may be the defining feature of malignancy in skin cancers. In addition, there have been other examples postulating that HGF secreted by stromal cells could facilitate invasion, according to c-Met expression in various c-Met over-expressing cancers, including gastric adenocarcinoma, colorectal carcinoma, thyroid cancer, head and neck cancer and breast cancers31-34. There are now substantial hypotheses that RTK such as c-Met may play a role in tumorigenesis and tumor progression, with its anti-apoptotic and pro-invasive activities helping to overcome the selective barriers to cancer progression17. Indeed, the HGF/c-Met system functions in a pro-angiogenic role; these activities have been known to induce tumor invasion and metastasis. This is supported by our study, where deeper MMs displayed higher expressions of c-Met.
Therefore, c-Met is thought to be closely involved in skin cancer development, especially MMs. Malignancy and prognosis can be presumed according to the level of c-Met expression in MMs.
Figures and Tables
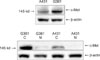 | Fig. 1The Immunoprecipitation and subcellular fraction Western blot assay in the immunoprecipitates. Based on immunoprecipitate analysis involving c-Met expression in A431 cells and G361 cells, c-Met expression was observed in both cell lines, but G361 cells were more strongly expressed. In a subcellular Western blot analysis of c-Met expression study, c-Met expression was found in the cytoplasm. A431: squamous cell carcinoma cell line, G361: malignant melanoma cell line, C: cytoplasm, N: nucleus. |
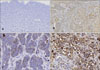 | Fig. 2Immunohistochemical analysis of c-Met expression in (A) normal skin tissue, (B) squamous cell carcinoma, (C) basal cell carcinoma, and (D) malignant melanoma. All malignant skin cancers displayed c-Met expression, but malignant melanoma was expressed more strongly (A: ×100, B, C, D: ×200). |
 | Fig. 3Immunohistochemical analysis of c-Met expression in malignant melanomas (MMs), squamous cell carcinomas (SCCs), basal cell carcinomas (BCCs), and normal human skin. c-Met was strongly expressed in malignant melanomas, while the c-Met was moderately expressed in SCCs and slightly expressed in BCCs. The c-Met was hardly expressed in normal human skin tissues. |
 | Fig. 4(A) Immunohistochemical analysis of c-Met expression in superficial malignant melanoma, and (B) deeper invasive malignant melanoma tissues. Malignant melanomas have stronger positive staining than other cancers. Indeed, deeper melanoma tissues which had increased breslow level melanomas are observed to have more increased positive staining to the immunohistochemicals (×400). |
References
1. Wolf HK, Zarnegar R, Michalopoulos GK. Localization of hepatocyte growth factor in human and rat tissues: an immunohistochemical study. Hepatology. 1991. 14:488–494.

2. Wang Y, Selden AC, Morgan N, Stamp GW, Hodgson HJ. Hepatocyte growth factor/scatter factor expression in human mammary epithelium. Am J Pathol. 1994. 144:675–682.
4. Peruzzi B, Bottaro DP. Targeting the c-Met signaling pathway in cancer. Clin Cancer Res. 2006. 12:3657–3660.

5. Pisters LL, Troncoso P, Zhau HE, Li W, von Eschenbach AC, Chung LW. c-Met proto-oncogene expression in benign and malignant human prostate tissues. J Urol. 1995. 154:293–298.

6. Matteucci E, Bendinelli P, Desiderio MA. Nuclear localization of active HGF receptor Met in aggressive MDA-MB231 breast carcinoma cells. Carcinogenesis. 2009. 30:937–945.

7. Gomes DA, Rodrigues MA, Leite MF, Gomez MV, Varnai P, Balla T, et al. c-Met must translocate to the nucleus to initiate calcium signals. J Biol Chem. 2008. 283:4344–4351.

8. Rong S, Jeffers M, Resau JH, Tsarfaty I, Oskarsson M, Vande Woude GF. Met expression and sarcoma tumorigenicity. Cancer Res. 1993. 53:5355–5360.
9. Prat M, Narsimhan RP, Crepaldi T, Nicotra MR, Natali PG, Comoglio PM. The receptor encoded by the human c-Met oncogene is expressed in hepatocytes, epithelial cells and solid tumors. Int J Cancer. 1991. 49:323–328.

10. Sheth PR, Watowich SJ. Biochemical ground-rules regulating c-Met receptor tyrosine kinase activation and signaling. Cancer Ther. 2006. 4:1–12.
11. Cooper CS, Park M, Blair DG, Tainsky MA, Huebner K, Croce CM, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984. 311:29–33.

12. Kuniyasu H, Yasui W, Yokozaki H, Kitadai Y, Tahara E. Aberrant expression of c-met mRNA in human gastric carcinomas. Int J Cancer. 1993. 55:72–75.

13. Natali PG, Nicotra MR, Di Renzo MF, Prat M, Bigotti A, Cavaliere R, et al. Expression of the c-Met/HGF receptor in human melanocytic neoplasms: demonstration of the relationship to malignant melanoma tumour progression. Br J Cancer. 1993. 68:746–750.

14. Di Renzo MF, Olivero M, Katsaros D, Crepaldi T, Gaglia P, Zola P, et al. Overexpression of the Met/HGF receptor in ovarian cancer. Int J Cancer. 1994. 58:658–662.
15. Boix L, Rosa JL, Ventura F, Castells A, Bruix J, Rodés J, et al. c-met mRNA overexpression in human hepatocellular carcinoma. Hepatology. 1994. 19:88–91.

16. Belfiore A, Gangemi P, Costantino A, Russo G, Santonocito GM, Ippolito O, et al. Negative/low expression of the Met/hepatocyte growth factor receptor identifies papillary thyroid carcinomas with high risk of distant metastases. J Clin Endocrinol Metab. 1997. 82:2322–2328.

17. Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov. 2008. 7:504–516.

18. Pisters LL, el-Naggar AK, Luo W, Malpica A, Lin SH. c-Met proto-oncogene expression in benign and malignant human renal tissues. J Urol. 1997. 158:724–728.

19. Natali PG, Prat M, Nicotra MR, Bigotti A, Olivero M, Comoglio PM, et al. Overexpression of the met/HGF receptor in renal cell carcinomas. Int J Cancer. 1996. 69:212–217.

20. Wagatsuma S, Konno R, Sato S, Yajima A. Tumor angiogenesis, hepatocyte growth factor, and c-Met expression in endometrial carcinoma. Cancer. 1998. 82:520–530.

21. Humphrey PA, Zhu X, Zarnegar R, Swanson PE, Ratliff TL, Vollmer RT, et al. Hepatocyte growth factor and its receptor (c-Met) in prostatic carcinoma. Am J Pathol. 1995. 147:386–396.
22. Ueki T, Fujimoto J, Suzuki T, Yamamoto H, Okamoto E. Expression of hepatocyte growth factor and its receptor c-met proto-oncogene in hepatocellular carcinoma. Hepatology. 1997. 25:862–866.

23. Berchuck A, Soisson AP, Clarke-Pearson DL, Soper JT, Boyer CM, Kinney RB, et al. Immunohistochemical expression of CA 125 in endometrial adenocarcinoma: correlation of antigen expression with metastatic potential. Cancer Res. 1989. 49:2091–2095.
24. Aasmundstad TA, Haugen OA, Johannesen E, Høe AL, Kvinnsland S. Oestrogen receptor analysis: correlation between enzyme immunoassay and immunohistochemical methods. J Clin Pathol. 1992. 45:125–129.

25. Ersoy B, Ozbilgin K, Kasirga E, Inan S, Coskun S, Tuglu I. Effect of growth hormone on small intestinal homeostasis relation to cellular mediators IGF-I and IGFBP-3. World J Gastroenterol. 2009. 15:5418–5424.

26. Ma PC, Maulik G, Christensen J, Salgia R. c-Met: structure, functions and potential for therapeutic inhibition. Cancer Metastasis Rev. 2003. 22:309–325.
27. Duh FM, Scherer SW, Tsui LC, Lerman MI, Zbar B, Schmidt L. Gene structure of the human MET proto-oncogene. Oncogene. 1997. 15:1583–1586.

28. Boccaccio C, Comoglio PM. Invasive growth: a Met-driven genetic programme for cancer and stem cells. Nat Rev Cancer. 2006. 6:637–645.

29. Rubin JS, Bottaro DP, Aaronson SA. Hepatocyte growth factor/scatter factor and its receptor, the c-met proto-oncogene product. Biochim Biophys Acta. 1993. 1155:357–371.

30. Chung LW, Li W, Gleave ME, Hsieh JT, Wu HC, Sikes RA, et al. Human prostate cancer model: roles of growth factors and extracellular matrices. J Cell Biochem Suppl. 1992. 16H:99–105.

31. Scarpino S, Stoppacciaro A, Colarossi C, Cancellario F, Marzullo A, Marchesi M, et al. Hepatocyte growth factor (HGF) stimulates tumour invasiveness in papillary carcinoma of the thyroid. J Pathol. 1999. 189:570–575.

32. Corso S, Migliore C, Ghiso E, De Rosa G, Comoglio PM, Giordano S. Silencing the MET oncogene leads to regression of experimental tumors and metastases. Oncogene. 2008. 27:684–693.

33. Beviglia L, Matsumoto K, Lin CS, Ziober BL, Kramer RH. Expression of the c-Met/HGF receptor in human breast carcinoma: correlation with tumor progression. Int J Cancer. 1997. 74:301–309.

34. Lamszus K, Jin L, Fuchs A, Shi E, Chowdhury S, Yao Y, et al. Scatter factor stimulates tumor growth and tumor angiogenesis in human breast cancers in the mammary fat pads of nude mice. Lab Invest. 1997. 76:339–353.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download