Abstract
Background
The characterization of progenitor/keratinocyte stem cells (KSC) remains an unachieved goal. A previous study showed that rapid adhering cells to collagen IV had the characteristics of putative progenitor/KSCs.
Objective
The purpose of this study was to investigate the genetic expression of rapid adhering cells compared to non adhering cells to determine the characteristic of KSCs.
Methods
We isolated rapid adhering cells representative of KSCs from non adhering cells representative of transient amplifying cells. In addition, we differentiated cells from human tonsilar keratinocytes utilizing the adhering capability of the KSCs to collagen IV. Annealing control primer based differentially displayed polymerase chain reaction (PCR) was performed as well as Western blot analysis.
Results
The levels of mitochondria-related gene expression were low in the rapid adhering cells compared to the non adhering cells. Mitochondrial complex I, COX IV, peroxiredoxins (I, II and IV) and mitochondrial membrane potential were all low in the rapid adhering cells compared to the non adhering cells.
Human keratinocytes are constantly renewed and replaced by a population of keratinocyte stem cells (KSC) located in the basal layer of the epidermis1-10. KSCs are responsible for the maintenance of skin cell homeostasis, and are believed to be resistant to noxious environmental stimuli; they give rise to fast-dividing transient amplifying cells (TAC) committed to terminal differentiation, while retaining their self-renewal capacity1-7. Although α6 integrin, β1 integrin, and p63 have been reported as markers for KSCs, FACS using these markers has not provided practical yields of viable KSCs6-10. An adhesion method has been developed using the rapid adhesiveness of KSCs to collagen type IV, fibronectin and extacellular matrix4-6,9,10 and has been assessed using many different methods of varying sensitivity, complexity and time4,5. Kim et al.9 in 2004, suggested that rapid adhering (R.A.) cells in the adhesion assay represented KSCs and therefore, this assay could be used to isolate KSCs. Our previous work using this adhesion assay10 showed that R.A. cells had the characteristics of KSCs including a small number of cells with undifferentiated morphology, and a stronger expression of α6 integrin, β1 integrin, β catenin and p6310,11. The goal of this study was to use the adhesion assay and annealing control primer based differentially displayed polymerase chain reaction (PCR) to further characterize the KSCs. We evaluated whether the adhesion assay showed a marked difference in some of the important characteristics of rapid adhering cells compared to non adhering cells by differentially expressed genes (DEGs)12,13. Subsequent Western blot analysis was used to confirm the differences observed.
Primary cultures with keratinocytes from three different tonsilar tissues were established. The keratinocytes were cultured in keratinocyte growth medium (Cambrex, Walkersville, MD, USA) as previously described14. The third passaged cells were placed in human collagen type IV (Sigma-Aldrich, St. Louis, MO, USA) coated dishes and incubated for 10 min as previously described10. Rapidly adhering (R.A.) cells9,15 were collected after vigorous washing and considered to be KSC fractions. After 90 min, suspended (non adhering, N.A.)9 cells were collected and considered to be TAC fractions that were post-mitotic differentiated cells10.
Total RNAs extracted from the R.A. and N.A. samples were reverse transcribed for 1.5 h at 42℃ in a final reaction volume of 20µl containing 3µg of the purified total RNA, 4µl of reaction buffer (Promega, Madison, WI, USA), 5µl of dNTPs (each 2 mM), 2µl of 10µM dT-ACP1 primer (5'-CTGTGAATGCTGCGACTACGATIIIIIT(18)-3'), 0.5µl of RNasin (RNase Inhibitor (40 U/µl; Promega), and 1µl of murine leukemia virus reverse transcriptase (200 U/µl; Promega). The DEGs were screened by the ACP-based PCR method12,13 using GeneFishing™ DEG kits (Seegene, Seoul, Korea). The PCR protocol for second-strand synthesis was one cycle at 94℃ for 1 min, followed by 50℃ for 3 min, and 72℃ for 1 min. The second-stage PCR amplification protocol was 40 cycles of 94℃ for 40 s, followed by 65℃ for 40 s, 72℃ for 40 s, followed by a 5 min final extension at 72℃. The amplified PCR products were separated on a 2% agarose gel stained with ethidium bromide. The differentially expressed bands were extracted and cloned into a TOPO TA cloning vector (Invitrogen, Carlsbad, CA, USA) and complete sequences were analyzed by searching for similarities using the BLASTX search program at the GenBank of the National Center for Biotechnology Information.
Total cell lysates (20 ug protein) form R.A. and N.A. cells underwent 8~16% SDS-PAGE and were transferred onto a PVDF membrane. To compare the differences in the expression of R.A and N.A. cells, other cells that had intermediate adherence ability were removed.
The DEGs, by ACP-based PCR, showed primarily mitochrondrial-related genes. Therefore, we investigated representative proteins related to mitochondrial biogenesis and to the respiratory chain including: mitochondrial complexes I to V, and peroxiredoxins 1 to 6. The primary antibodies included antibodies against mitochondrial complex I (NADH ubiquinone oxicoreductase 39 kDa a subcomplex 9, NDUFA9), complex II (Succinate-ubiquinone oxicoreductase 70 kDa flavoprotein, SDHA), complex III (Ubiquinone-cytochrome c, core II, UQCRC2), complex IV (cytochrome C oxidase subunit I, COXI and, subunit IV, COXIV) and complex V (ATP synthase, F1 complex a, ATP5A1) (all Invitrogen). In addition, antibodies against peroxiredoxins 1~6 (Prx 1~6) (Lab Frontier, Seoul, Korea), S-100 calcium binding protein A8 (Santa Cruz Bio, Santa Cruz, CA, USA) and β-actin (Sigma-Aldrich) were evaluated against the loading of proteins. The membrane was incubated with goat anti-mouse IgG or goat anti-rabbit IgG conjugated with horseradish peroxidase (Santa Cruz Bio). The immunoblots were developed by the ECL reaction (Amersham, Piscataway, NJ, USA).
R.A. and N.A. cells were stained with 150 nM TMRE (Invitrogen) in the dark at 37℃ for 30 min. The cells were then washed, resuspended in phosphophate-buffered saline containing 15 nM TMRE, and analyzed by flow cytometry (FACS Calibur, BD Biosciences, Franklin lakes, NJ, USA). For the mitotracker staining, the cells were cultured in chamber slides overnight, then washed and fixed with 3.7% paraformaldehyde (Sigma-Aldrich). The cells were stained with 500 nM of MitoTracker Orange CMTMRos (Invitrogen) in the dark at room temperature for 20 min after permeabilization with triton X-100 (Sigma-Aldrich). The fluorescent signal was visualized by a laser-scanning confocal microscope (LSM510, Germany).
The products separated for the DEGs on agarose gels showed 10 different cDNA bands (C1-C10) by hatched blastocyst-specific expression (Table 1). The 10 different DEG clone found by comparing the bands of the R.A. and N.A. cells included four mitochondrion-related genes: C1 (mitochondrion), C2 (mitochondrion), C5 (mitochondrion) and C8 (mitochondrial DNA). In addition, C7 (S100 calcium binding protein A8) and C9 (nitric oxide synthase interacting protein) also differed in the DEG clones. However, C6 was not significantly different by sequence comparison.
Western blot analysis showed that S100 calcium binding protein A8 showed no difference in the protein expression when R.A. and N.A. cells were compared. Western blot analysis using antibodies associated with mitochondrial biogenesis showed that in all the samples with R.A. cells, the nuclear DNA (nDNA) encoded mitochondrial respiratory complex subunits; NDUFA9 of complex I and COX IV of complex IV were reduced compared to the N.A. cells (Fig. 1A). The Mitochondrial DNA (mtDNA)-encoded COX I subunit of complex IV was reduced or equivocal. Interestingly other nDNA-encoded subunits of complex II (SDHA), III (UQCRC2), and V (ATP5A1) did not differ in their expression in repeated experiments. The Western blot analysis of peroxiredoxin isoforms showed that Prx 1, Prx 2, and Prx 4 were consistently reduced compared to the N.A. cells; however, the Prx 3 and Prx 5 expression did not differ (Fig. 1B).
We isolated R.A. cells representative of a KSC enriched population and N.A. cells representative of TAC and differentiated cells. We used ACP-based PCR technology to identify DEGs between the R.A. and N.A. cells. ACP technology is based on the unique structure of a specific ACP which contains distinct 3'- and 5'- end regions separated by a regulator, and the interaction of each portion of this primer during two-stage PCR12,13. This system facilitates the identification of DEGs from small samples without generating false positive results13.
The results of the experiments showed differences between R.A. and N.A. cells in mitochondria-related gene expression. The results showed that mitochondrial complex I, COX IV, Prx 1, Prx 2, Prx 4 and the mitochondrial membrane potential were low in the R.A. cells compared to the N.A. cells. These consistent differences confirmed that this adhesion assay is useful for the isolation of KSCs.
Mitochondria are the major generators of cellular adenosine tri-phosphate (ATP) through oxidative phosphorylation16,17. In addition, mitochondria are critical to both apoptosis and necrosis17. The resistance of KSC's to damaging environmental stimuli10 may in part be due to the low activity of mitochondria. Similarly, human hematopoietic stem cells have low amounts of mitochondrial respiratory chain complexes and poor oxidative phosphorylation16. The Low activity of mitochondria in R.A. cells may be related to low ROS generation in KSCs. Induction of mitogenic signaling causes the formation of ROS, which can also cause mitogenic stimulation18. Thus low function or less differentiation of mitochondria may make KSCs, under stable conditions, divide slowly. In addition, mitochondrial dysfunction has been reported in human colon stem cells19. Furthermore, another report showed that pathogenic mutations in the mitochondrial genome contributed to the promotion of cancer by preventing apoptosis20; stem cells share with cancer cells the ability to escape from apoptosis21.
The results of this study showed that Prx 1, Prx 2, and Prx 4 were low in the KSCs compared to the TACs. Prx is known to protect cells and tissues from oxidative damage by removing toxic hydrogen peroxide18,22,23. Previously, Prx 1 and Prx 2 were found to have low levels of expression in undifferentiated human embryonic stem cells (ESCs) and increased levels in differentiated cells23-25. The role of Prx 4 is not well known. A prior study showed that the mitochondrial mass was almost absent in the undifferentiated ESCs and dramatically appeared as differentiation progressed23. In our study, a functional decrement, rather than a decreased amount, of mitochondria in the R.A. cells was observed; since mitotracker staining was stained regardless of the membrane potential. Reduction of complex I, a coordinator of other complexes25-27, in the R.A. cells, suggests that the general mitochondrial complex activity was decreased in the KSC fraction. Complex IV includes the mitochondria-encoded enzymes, COX subunit I and subunit IV27; therefore, reduction of components of complex IV in the R.A. cells may also support the low mitochondrial activity in the KSCs.
In comparison to the N.A. cell population, the R.A. cells exhibit lower levels of aerobic respiration-related proteins. R.A. cells may be less susceptible to oxidative damage. Taken together, our results suggest that reduced mitochondrial biogenesis may be a characteristic of KSCs. However, additional studies are needed to confirm this possibility.
Figures and Tables
Fig. 1
Mitochondrial function in the R.A. and N.A. cells. Primary cultures of oral tonsilar mucosal keratinocytes were established and separated by their different adhesion to collagen type IV. The expression of mitochondrial proteins in R.A. and N.A. cells was confirmed by Western blot analysis. (A) The expression of mitochondrial complexes in R.A. and N.A. cells. (B) The expression of peroxiredoxin 1~6 in R.A. and N.A. cells.

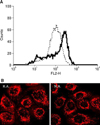
Fig. 2
TMRE flow cytometry and confocal microscopy for mitochondria. (A) R.A. (dotted line) and N.A. (solid line) cells were stained with TMRE and analyzed by FACS. The abscissa shows the intensity of the fluorescence, the ordinate the relative number of cells. (B) R.A. and N.A. cells were treated with MitoTracker Orange CMTMRos to stain the mitochondria and then studies under a confocal microscope.
References
1. Potten CS. Keratinocyte stem cells, label-retaining cells and possible genome protection mechanisms. J Investig Dermatol Symp Proc. 2004. 9:183–195.

2. Pellegrini G, Bondanza S, Guerra L, De Luca M. Cultivation of human keratinocyte stem cells: current and future clinical applications. Med Biol Eng Comput. 1998. 36:778–790.

3. Kaur P. Interfollicular epidermal stem cells: identification, challenges, potential. J Invest Dermatol. 2006. 126:1450–1458.

4. Kaur P, Li A. Adhesive properties of human basal epidermal cells: an analysis of keratinocyte stem cells, transit amplifying cells, and postmitotic differentiating cells. J Invest Dermatol. 2000. 114:413–420.

5. Potten CS, Booth C. Keratinocyte stem cells: a commentary. J Invest Dermatol. 2002. 119:888–899.
6. Jones PH, Watt FM. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell. 1993. 73:713–724.

7. Tani H, Morris RJ, Kaur P. Enrichment for murine keratinocyte stem cells based on cell surface phenotype. Proc Natl Acad Sci U S A. 2000. 97:10960–10965.

8. Jones PH, Harper S, Watt FM. Stem cell patterning and fate in human epidermis. Cell. 1995. 80:83–93.

9. Kim DS, Cho HJ, Choi HR, Kwon SB, Park KC. Isolation of human epidermal stem cells by adherence and the reconstruction of skin equivalents. Cell Mol Life Sci. 2004. 61:2774–2781.

10. Jeong EJ, Choi SH, Chang SE, Chang HW, Roh JL, Lee SW, et al. Putative progenitor/stem cells isolated from human oral mucosa are resistant to ionizing radiation. J Dermatol Sci. 2008. 50:65–68.

11. Radu E, Simionescu O, Regalia T, Dumitrescu D, Popescu LM. Stem cells (p63(+)) in keratinocyte cultures from human adult skin. J Cell Mol Med. 2002. 6:593–598.

12. Li S, Ross DT, Kadin ME, Brown PO, Wasik MA. Comparative genome-scale analysis of gene expression profiles in T cell lymphoma cells during malignant progression using a complementary DNA microarray. Am J Pathol. 2001. 158:1231–1237.

13. Hwang KC, Cui XS, Park SP, Shin MR, Park SY, Kim EY, et al. Identification of differentially regulated genes in bovine blastocysts using an annealing control primer system. Mol Reprod Dev. 2004. 69:43–51.

14. Hong KK, Lew BL, Kim YI, Lee JW, Kim NI. The effect of TNF-alpha and IFN-gamma on the telomerase activity of cultured human keratinocytes. Ann Dermatol. 2007. 19:147–152.

15. Dong R, Liu X, Liu Y, Deng Z, Nie X, Wang X, et al. Enrichment of epidermal stem cells by rapid adherence and analysis of the reciprocal interaction of epidermal stem cells with neighboring cells using an organotypic system. Cell Biol Int. 2007. 31:733–740.

16. Piccoli C, Ria R, Scrima R, Cela O, D'Aprile A, Boffoli D, et al. Characterization of mitochondrial and extra-mitochondrial oxygen consuming reactions in human hematopoietic stem cells. Novel evidence of the occurrence of NAD(P)H oxidase activity. J Biol Chem. 2005. 280:26467–26476.

17. Kim JY, Kim YH, Chang I, Kim S, Pak YK, Oh BH, et al. Resistance of mitochondrial DNA-deficient cells to TRAIL: role of Bax in TRAIL-induced apoptosis. Oncogene. 2002. 21:3139–3148.

18. Lee JB, Yun SJ, Chae HZ, Won YH, Kim YP, Lee SC. Expression of peroxiredoxin and thioredoxin in dermatological disorders. Br J Dermatol. 2002. 146:710–712.

19. Greaves LC, Preston SL, Tadrous PJ, Taylor RW, Barron MJ, Oukrif D, et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc Natl Acad Sci U S A. 2006. 103:714–719.

20. Shidara Y, Yamagata K, Kanamori T, Nakano K, Kwong JQ, Manfredi G, et al. Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res. 2005. 65:1655–1663.

21. Finlan LE, Hupp TR. Epidermal stem cells and cancer stem cells: insights into cancer and potential therapeutic strategies. Eur J Cancer. 2006. 42:1283–1292.

22. Lee SC, Chae HZ, Lee JE, Kwon BD, Lee JB, Won YH, et al. Peroxiredoxin is ubiquitously expressed in rat skin: isotype-specific expression in the epidermis and hair follicle. J Invest Dermatol. 2000. 115:1108–1114.

23. Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park DJ, et al. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem Biophys Res Commun. 2006. 348:1472–1478.

24. Lonergan T, Brenner C, Bavister B. Differentiation-related changes in mitochondrial properties as indicators of stem cell competence. J Cell Physiol. 2006. 208:149–153.

25. St John JC, Ramalho-Santos J, Gray HL, Petrosko P, Rawe VY, Navara CS, et al. The expression of mitochondrial DNA transcription factors during early cardiomyocyte in vitro differentiation from human embryonic stem cells. Cloning Stem Cells. 2005. 7:141–153.
26. St John JC, Amaral A, Bowles E, Oliveira JF, Lloyd R, Freitas M, et al. The analysis of mitochondria and mitochondrial DNA in human embryonic stem cells. Methods Mol Biol. 2006. 331:347–374.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download