Abstract
Background
Malassezia yeasts are normal flora of the skin found in 75~98% of healthy subjects. The accurate identification of the Malassezia species is important for determining the pathogenesis of the Malassezia yeasts with regard to various skin diseases such as Malassezia folliculitis, seborrheic dermatitis, and atopic dermatitis.
Objective
This research was conducted to determine a more accurate and rapid molecular test for the identification and classification of Malassezia yeasts.
Methods
We compared the accuracy and efficacy of restriction fragment length polymorphism (RFLP) and the nested polymerase chain reaction (PCR) for the identification of Malassezia yeasts.
Results
Although both methods demonstrated rapid and reliable results with regard to identification, the nested PCR method was faster. However, 7 different Malassezia species (1.2%) were identified by the nested PCR compared to the RFLP method.
Conclusion
Our results show that RFLP method was relatively more accurate and reliable for the detection of various Malassezia species compared to the nested PCR. But, in the aspect of simplicity and time saving, the latter method has its own advantages. In addition, the 26S rDNA, which was targeted in this study, contains highly conserved base sequences and enough sequence variation for inter-species identification of Malassezia yeasts.
The Malassezia species are considered part of the normal flora of the skin and are associated with pityriasis versicolor, Malassezia folliculitis, seborrheic dermatitis, dandruff, atopic dermatitis and psoriasis1. In 1996 Gueho et al.2 classified them into seven species: M. furfur, M. pachydermatis, M. sympodialis, M. globosa, M. obtusa, M. restricta and M. slooffiae. Recently, on the basis of common DNA sequences and molecular biology testing methods, four new species have been added: M. dermaits, M. japonica, M. nana and M. yamatoensis3-7.
Many investigators involved in Malassezia yeast research still use the morphological analysis of size, surface, color, and shape of the cultured colony and biochemical analysis for identification of this organism8-11. However, the morphological analysis is usually a time-consuming, multi-step process necessitating several experimental techniques; in addition, this approach does not take the taxonomic component into consideration, and thus the genetic link between species cannot be determined by conventional methods. Therefore, the morphological and biochemical methods currently used for the study of this yeast limit the identification and classification of new species.
To overcome the limits of morphological assessment, recent studies have used a variety of molecular methods such as the nested polymerase chain reaction (PCR)12, real-time PCR13, pulsed field gel electrophoresis (PFGE)14, amplified fragment length polymorphism (AFLP)15,16, denaturing gradient gel electrophoresis (DGGE)16, random amplification of polymorphic DNA (RAPD)16,17, single strand conformation polymorphism (SSCP)18, terminal fragment length polymorphism (tFLP)19, restriction fragment length polymorphism (RFLP)20-23, and sequencing analysis24.
To investigate a more accurate and rapid molecular approach to the identification and classification of Malassezia yeasts, we compared the accuracy and efficacy of the RFLP and nested PCR methods.
Normal subjects included 110 healthy volunteers (60 males, 50 females); 0~80 years of age without any dermatoses of the examined regions. Sterile cotton swabs were moistened with wash fluid containing 0.1% Triton X-100 in 0.075 M phosphate buffer (pH 7.9), and rubbed gently, with rotation on the skin. Swabbing was performed for 10 sec on sites including: the scalp, forehead, cheek, chest, and thighs. The swabs were immediately placed in Leeming and Notman agar media. The investigations were conducted according to the Declaration of Helsinki Principles. Written informed consent was obtained from each subject before the procedure.
The yeast was cultured on agar plates with Leeming and Notman agar (LNA) - glycerol monoesterate (BDH, Poole, UK) 0.5 g, bacteriological peptone (Oxoid, Hampshire, UK) 20 g, glucose (Oxoid, Hampshire, UK) 5 g, yeast extract (Oxoid, Hampshire, UK) 0.1 g, ox bile (Merck, Darmstadt, Germany) 4 g, agar No.1 (Oxoid, Hampshire, UK) 12 g, Tween 60 (Yakuri, Osaka, Japan) 0.5 ml, glycerol (Tedia, Fairfield, USA) 1 ml, 10 ml of whole-fat cow's milk per liter that contained cycloheximide (Sigma, St Louis, MO, USA) 200 mg, and chloramphenicol (Sigma, St Louis, MO, USA) 50 mg. The plates were incubated at 34℃ for 14 days.
The yeasts grown in agar were harvested and resuspended in 0.4 ml of lysis buffer (100 mM Tris-HCl pH 8.0, 1.0% SDS, 2.0% Triton X-100, 10 mM EDTA, 100 mM NaCl). Equal volumes of phenol/chloroform/isoamyl alcohol (phenol: chloroform: isoamyl alcohol=25:24:1, v/v) and glass beads (0.5 mm) were added, and the mixture was vortexed for 10 min. The samples were centrifuged at 12,000×g for 15 min. Total DNA was precipitated from the aqueous fraction with isopropanol and centrifuged at 12,000×g for 20 min at 4℃. The DNA pellet was washed in 70% ethanol and resuspended in sterile water.
To amplify 26S rDNA from genomic DNA, the reaction mixture contained 25 mM of each dNTP, 10X PCR buffer, 5X Q buffer, 0.5 µM primer, 0.4 µm forward primer (5'-TAACAAGGATTCCCCTAGTA-3'), reverse primer (5'-ATTACGCCAGCATCCTAAG-3'), and 1.25 U Hot StarTaq polymerase in 50 µl reaction volume. Thirty five cycles with the following protocol were programmed: denaturation for 45 sec at 94℃; annealing for 45 sec 55℃; extension for 1 min at 72℃. After the confirmation of amplified 26S rDNA, the PCR products were purified using an Accu-Prep PCR purification kit (Bioneer, Daejeon, Korea). The absorbance at 260 nm and the 260/280 ratio (as a measure of DNA quality) were obtained for each purified PCR product sample using the NanoDrop spectrophotometer ND-1000 (NanoDrop Technologies) and the estimated concentration of DNA (ng/µl). The 26S rDNA was confirmed by running 50 ng of 26S rDNA on a 2% agarose gel. Two restriction enzymes, Hha1 (Takara Biomedicals, Otsu, Japan) and BstF51 (SibEnzyme, Novosibirsk, Russia) were used to perform the 26S rDNA-RFLP study of Malassezia. In this experiment, the restriction enzyme digestion was performed with 10 X PCR buffer, 10 U of the restriction enzyme, and PCR products 7.5 µl, of each sample was examined, from which the maximum expected concentration was 50 ng/µl (1 µg DNA in 20 µl recovery solution), which added up to 20 µl. After the reaction at 37℃ for 3 hours, the electrophoresis was performed with a 3.5% (w/v) NuSieve GTG agarose gel (FMC, Rockland, ME, USA) with 100 volts and stained with ethidium bromide. The restriction fragments were analyzed by the size and number of the DNA fragments under UV transillumination (Fig. 1).
To amplify rDNA from genomic DNA, a nested PCR was carried out using primers for Malassezia. The first amplification mix was carried out in a 20 µl reaction consisting of 2× pre mix (2 µl 10× buffer, 1.6 µl of 25 mM MgCl2, 0.25 µl of 10 mM dNTPs, nTaq-Hot DNA polymerase -Enzynomics-Korea), 1 µl of 2.5 µM primer and 10 ng of purified DNA. Primer pairs selecting for the Malassezia nana genes for ITS1, 5.8S rRNA, ITS2 are shown after an initial 5 min denaturation at 95℃, 20 cycles of 95℃ for 30 sec, 53.5℃ for 30 sec and 72℃ for 1 min were carried out, followed by a 7 min extension at 72℃ using a thermal cycler (Gene Amp PCR System 2400-Applied Biosystem, Monza, Italy). The reaction mixture for the second amplification round was the same as for the first, except for the "inner" primers [forward primer (5-GTCTCTGGCGCCTAACCCACTATA -3), reverse primer (5-TCCACGTACATACAACCATACGA-3)] were used instead of the "outer" primers [forward primer (5-GCCATACGGACGATAA-3), reverse primer (5-AGCAAATGACGTATCATGCCATGC-3)]. For the second amplification round 49 µl of amplification mix and 1 µl of the first amplification round PCR product were used. The thermal cycling was repeated as for the first amplification round using 30 cycles after the initial 2 min denaturation. Each amplification run contained a negative control, consisting of water and a positive control. Analysis of the PCR products was performed by 2% agarose gel electrophoresis followed by visualization with ethidium bromide (0.5 µg/ml) staining and UV illumination to confirm the expected products (Fig. 1).
The overall positive culture rate of the Malassezia yeasts samples from different body sites of 110 persons was 63.6%, with 350 positive samples out of 550 samples.
Using 26S rDNA PCR-RFLP analysis, we successfully identified 11 species of Malassezia yeasts from the standard Malassezia species (Fig. 2). In addition, we identified six species of Malassezia yeasts from 350 positive samples. The results showed that M. globosa was identified most frequently in 116 samples (33.1%); M. restricta in 106 samples (30.3%); M. sympodialis in 55 samples (15.6%); M. furfur in 39 samples (11.1%); M. slooffiae in 5 samples (1.4%), and M. dermatitis in 6 samples (1.6%). As for the 23 samples with co-identification of more than two Malassezia species, they were excluded from the analysis for a more direct comparison between the RFLP and nested PCR methods (Table 1).
Using nested PCR analysis, we successfully identified 11 species of Malassezia yeasts from the standard Malassezia species (Fig. 3). In addition, 6 species of Malassezia yeasts were identified from 327 positive samples that were already confirmed by RFLP methods. These results showed that M. globosa was identified most frequently in 111 samples (31.7%); M. restricta in 105 samples (30.0%); M. sympodialis in 54 samples (15.4%); M. furfur in 39 samples (11.1%); M. slooffiae in 5 samples (1.4%), and M. dermatitis in 6 samples (1.7%) (Table 1).
The results of re-identifying and classifying Malassezia species using nested PCR among the 327 positive samples, where cases with two or more yeasts identified were excluded, showed that 320 out of 327 (98.8%) samples were identical with the results of the RFLP method. In cases with M. globosa, 111 out of 116 samples were identical, showing a 95.6% concordance rate, and among the 5 non-identical samples, 3 were later identified as M. obtusa (131 bp) and 2 as M. nana (114 bp) by the nested PCR. In addition, 105 out of 106 M. restricta samples identified by the RFLP method were identical, and the other 1 case was confirmed as M. obtusa (131 bp) by the nested PCR. As for M. sympodialis, 54 out of 55 samples were identical, and the other 1 case was confirmed to be M. nana (114 bp) by the nested PCR (Table 1).
Malassezia yeasts are normal flora found on the skin of 75~98% of healthy persons, and has also been cited as a causative organism in pityriasis versicolor and as an aggravating factor in various skin diseases, including atopic dermatitis and seborrheic dermatitis1. Especially in atopic dermatitis, Malassezia yeast act as an allergenic aggravating factor rather than as infectious agents. Among the Malassezia species, sensitization to M. sympodialis is highly specific for patients with atopic eczema and does not occur in patients with only inhalant allergies, urticaria, or allergic contact dermatitis25. Therefore, the accurate identification of the Malassezia species is important to determine the pathogenesis of Malassezia yeasts in a variety of skin diseases.
Many different morphological and molecular methods have been used for the identification and classification of Malassezia yeasts. Most prior studies have used morphological and biochemical methods of analysis, which are time-consuming and subject to controversy, due to ambiguous objective criteria. Among the molecular methods, each method has different diagnostic accuracy for identifying Malassezia yeasts, and can also be inaccurate when used for the investigation of Malassezia yeast pathogenesis. Therefore, it is important to ascertain the efficacy of molecular methods including accuracy and cost-efficacy.
In this study, we compared the accuracy and efficacy of RFLP and nested PCR, both relatively accurate methods for the identification of Malassezia yeasts. We already reported that 26S rDNA RFLP method was a sensitive and rapid method for identification system for Malassezia species, which showed more than 99% consistent rate with genebank homology of Malassezia yeasts when analyzing clinical isolates and performing 26S rDNA sequencing24. In the results of this study, although both methods showed relatively rapid and reliable results for identification, nested PCR method showed faster and time saving advantages, but some additional different Malassezia species (7 species: 1.2%) were identified by the the nested PCR compared to the RFLP method. This can be explained by the fact that the nested PCR resulted in a single band differences in the electrophoresis of the Malassezia species from genomic DNA, which may have biased the analysis because of small differences among single band patterns. On the other hand, the RFLP method showed multiple band differences in the electrophoresis, which could help with a more precise identification of Malassezia species compared to the nested PCR method.
The RFLP method used in this study enables the examination of genetic variations by cleaving the amplified DNA with restriction enzymes and analyzing the patterns of the fragments. The improved accuracy and rapid diagnosis make this a desirable approach. The nested PCR has the advantages of simplicity and rapid turn around for results, but may be less accurate.
The 26S rDNA, which was targeted in this study, contains highly conserved base sequences and enough sequence variation for inter-specific identification. In addition, it is compatible with morphological methods and appropriate for the identification currently known Malassezia species; it requires only two restriction enzymes, Hha I, BtsC I, and has been proven to be technically easier to perform than other molecular techniques.
Our results show that RFLP method was relatively more accurate and reliable for the detection of various Malassezia species than the nested PCR method. But, in the aspect of simplicity and time saving, the nested PCR method has its own advantages. New, more rapid and precise, molecular methods for the identification and classification of Malassezia yeasts are needed; further studies will likely involve the quantitative analysis of Malassezia microflora using a real-time PCR assay.
Figures and Tables
 | Fig. 2PCR-RFLP patterns of 26S rDNA PCR digested with Hha I (A), BtsC I (B) of 11 Malassezia standard strains. Lanes: M. molecular marker; 1. M. furfur (KCTC 7743); 2. M. sympodialis (KCTC 7985); 3. M. globosa (CBS 7966); 4. M. restricta (KCTC 7848); 5. M. slooffiae (KCTC 17431); 6. M. pachydermatis (KCTC 17008); 7. M. japonica (CBS 9432); 8. M. nana (JCM 12085); 9. M. dermatis (JCM 11348); 10. M. obtusa (KCTC 7847); 11. M. yamatoensis (CBS 9725). |
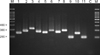 | Fig. 3Nested PCR products of standard Malassezia species. Lanes: M. molecular marker; 1. M. dermatis (JCM 11348); 2. M. furfur (KCTC 7743); 3. M. globosa (CBS 7966); 4. M. japonica (CBS 9432); 5. M. nana (JCM 12085); 6. M. obtuse (KCTC 7847); 7. M. pachydermatis (KCTC 17008); 8. M. restricta (KCTC 7848); 9. M. slooffiae (KCTC 17431); 10. M. sympodialis (KCTC 7985); 11. M. yamatoensis (CBS 9725); C. negative control. |
References
1. Gupta AK, Batra R, Bluhm R, Boekhout T, Dawson TL Jr. Skin diseases associated with Malassezia species. J Am Acad Dermatol. 2004. 51:785–798.
2. Gueho E, Midgley G, Guillot J. The genus Malassezia with description of four new species. Antonie Van Leeuwenhoek. 1996. 69:337–355.
3. Sugita T, Kodama M, Saito M, Ito T, Kato Y, Tsuboi R, et al. Sequence diversity of the intergenic spacer region of the rRNA gene of Malassezia globosa colonizing the skin of patients with atopic dermatitis and healthy individuals. J Clin Microbiol. 2003. 41:3022–3027.

4. Sugita T, Takashima M, Shinoda T, Suto H, Unno T, Tsuboi R, et al. New yeast species, Malassezia dermatis, isolated from patients with atopic dermatitis. J Clin Microbiol. 2002. 40:1363–1367.

5. Sugita T, Takashima M, Kodama M, Tsuboi R, Nishikawa A. Description of a new yeast species, Malassezia japonica, and its detection in patients with atopic dermatitis and healthy subjects. J Clin Microbiol. 2003. 41:4695–4699.

6. Hirai A, Kano R, Makimura K, Duarte ER, Hamdan JS, Lachance MA, et al. Malassezia nana sp. nov., a novel lipid-dependent yeast species isolated from animals. Int J Syst Evol Microbiol. 2004. 54:623–627.

7. Sugita T, Tajima M, Takashima M, Amaya M, Saito M, Tsuboi R, et al. A new yeast, Malassezia yamatoensis, isolated from a patient with seborrheic dermatitis, and its distribution in patients and healthy subjects. Microbiol Immunol. 2004. 48:579–583.

8. Lee YW, Yim SM, Lim SH, Choe YB, Ahn KJ. Quantitative investigation on the distribution of Malassezia species on healthy human skin in Korea. Mycoses. 2006. 49:405–410.

9. Guillot J, Gueho E, Lesourd M, Midgley G, Chevrier G, Dupont B. Identification of Malassezia species. A practical approach. J Mycol Med. 1996. 6:103–110.
10. Rodriguez-Valero S, Mesa LM, Gonzalez-Moran E, Delmonte ML, Robertiz S, Valero A. Phenotypic characterization of species of Malassezia in healthy skin of an university student population. Invest Clin. 2005. 46:329–335.
11. Bernier V, Weill FX, Hirigoyen V, Elleau C, Feyler A, Labreze C, et al. Skin colonization by Malassezia species in neonates: a prospective study and relationship with neonatal cephalic pustulosis. Arch Dermatol. 2002. 138:215–218.
12. Morishita N, Sei Y, Sugita T. Molecular analysis of Malassezia microflora from patients with pityriasis versicolor. Mycopathologia. 2006. 161:61–65.

13. Sugita T, Tajima M, Tsubuku H, Tsuboi R, Nishikawa A. Quantitative analysis of cutaneous Malassezia in atopic dermatitis patients using real-time PCR. Microbiol Immunol. 2006. 50:549–552.

14. Senczek D, Siesenop U, Bohm KH. Characterization of Malassezia species by means of phenotypic characteristics and detection of electrophoretic karyotypes by pulsed-field gel electrophoresis (PFGE). Mycoses. 1999. 42:409–414.
15. Gupta AK, Boekhout T, Theelen B, Summerbell R, Batra R. Identification and typing of Malassezia species by amplified fragment length polymorphism and sequence analyses of the internal transcribed spacer and large-subunit regions of ribosomal DNA. J Clin Microbiol. 2004. 42:4253–4260.

16. Theelen B, Silvestri M, Gueho E, van Belkum A, Boekhout T. Identification and typing of Malassezia yeasts using amplified fragment length polymorphism (AFLP), random amplified polymorphic DNA (RAPD) and denaturing gradient gel electrophoresis (DGGE). FEMS Yeast Res. 2001. 1:79–86.

17. Gandra RF, Simao RC, Matsumoto FE, da Silva BC, Ruiz LS, da Silva EG, et al. Genotyping by RAPD-PCR analyses of Malassezia furfur strains from pityriasis versicolor and seborrhoeic dermatitis patients. Mycopathologia. 2006. 162:273–280.

18. Gaitanis G, Velegraki A, Alexopoulos EC, Chasapi V, Tsigonia A, Katsambas A. Distribution of Malassezia species in pityriasis versicolor and seborrhoeic dermatitis in Greece. Typing of the major pityriasis versicolor isolate M. globosa. Br J Dermatol. 2006. 154:854–859.

19. Gemmer CM, DeAngelis YM, Theelen B, Boekhout T, Dawson TL Jr. Fast, noninvasive method for molecular detection and differentiation of Malassezia yeast species on human skin and application of the method to dandruff microbiology. J Clin Microbiol. 2002. 40:3350–3357.

20. Guillot J, Deville M, Berthelemy M, Provost F, Gueho E. A single PCR-restriction endonuclease analysis for rapid identification of Malassezia species. Lett Appl Microbiol. 2000. 31:400–403.

21. Makimura K, Tamura Y, Kudo M, Uchida K, Saito H, Yamaguchi H. Species identification and strain typing of Malassezia species stock strains and clinical isolates based on the DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J Med Microbiol. 2000. 49:29–35.

22. Gaitanis G, Velegraki A, Frangoulis E, Mitroussia A, Tsigonia A, Tzimogianni A, et al. Identification of Malassezia species from patient skin scales by PCR-RFLP. Clin Microbiol Infect. 2002. 8:162–173.

23. Mirhendi H, Makimura K, Zomorodian K, Yamada T, Sugita T, Yamaguchi H. A simple PCR-RFLP method for identification and differentiation of 11 Malassezia species. J Microbiol Methods. 2005. 61:281–284.

24. Lee YW, Lim SH, Ahn KJ. The application of 26S rDNA PCR-RFLP in the identification and classification of Malassezia yeast. Korean J Med Mycol. 2006. 11:141–153.
25. Casagrande BF, Fluckiger S, Linder MT, Johansson C, Scheynius A, Crameri R, et al. Sensitization to the yeast Malassezia sympodialis is specific for extrinsic and intrinsic atopic eczema. J Invest Dermatol. 2006. 126:2414–2421.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download