Abstract
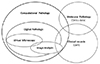
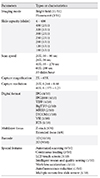
Pathology has a long history of artificial intelligence (AI) as much as any other field of medicine, and has used AI algorithms continuously. However, in Korea, pathology AI is unfamiliar even to the pathologists. In this article, I will summarize the terms and definitions, the basic elements of pathology AI, and the future direction. Digital pathology is a system or environment that digitizes glass slides into binary files, observes them through a monitor or any digital devices, interprets it, analyzes it, and maintains it. Computational pathology is a comprehensive concept of diagnosis support or research system that deals with image, text and omics data. Virtual microscopy is a method or technology that allows pathologists to view and share glass slides images from whole slide scanners. Image analysis is a technique or method that processes various digital images and quantifies features. The basic elements of pathology AI are as follows: environmental factors called digital pathology and technical elements such as AI, machine learning, and deep learning. Digital pathology workflow consists of three elements; acquisition or collection of data, data processing and data storage. The basic process of image analysis consists of preprocessing of image, identification of region of interest, and feature extraction. There is enormous potential for improvement of patient care through digital pathology and/or AI, and a harmonized discussion about activation of Korean digital pathology among government, academia and industry will be mandatory for future medicine and healthcare in Korea.
Figures and Tables
References
1. Borowiec S. AlphaGo seals 4-1 victory over Go grandmaster Lee Sedol. The Guardian. 2016. 03. 15.
2. Byford S. AlphaGo retires from competitive Go after defeating world number one 3-0. The Verge. 2017. 05. 27.
3. Fitzgerald J. IBM, NYC hospital training Watson supercomputer in cancer. Phys Org. 2012. 03. 22.
4. Granter SR, Beck AH, Papke DJ Jr. AlphaGo, Deep Learning, and the Future of the Human Microscopist. Arch Pathol Lab Med. 2017; 141:619–621.

5. Sharma G, Carter A. Artificial Intelligence and the Pathologist: Future Frenemies? Arch Pathol Lab Med. 2017; 141:622–623.

6. Granter SR, Beck AH, Papke DJ Jr. Straw Men, Deep Learning, and the Future of the Human Microscopist: Response to “Artificial Intelligence and the Pathologist: Future Frenemies”. Arch Pathol Lab Med. 2017; 141:624.

8. Louis DN, Gerber GK, Baron JM, Bry L, Dighe AS, Getz G, et al. Computational Pathology. An Emerging Definition. Arch Pathol Lab Med. 2014; 138:1133–1138.

10. Song SY. Artificial Intelligence in Pathology. In : The 25th federation meeting of Korean basic medical scientists 2017; 2017. p. S33.
11. Cucoranu IC, Parwani AV, Vepa S, Weinstein RS, Pantanowitz L. Digital pathology: A systematic evaluation of the patent landscape. J Pathol Inform. 2014; 5(1):16.

12. Dunn BE, Almagro UA, Choi H, Sheth NK, Arnold JS, Recla DL, et al. Dynamic-robotic telepathology: Department of Veterans Affairs feasibility study. Hum Pathol. 1997; 28:8–12.

13. Ferreirat R, Moon B, Humphriest J, Sussman A, Saltz J, Miller R, et al. The Virtual Microscope. Proc AMIA Annu Fall Symp. 1997; 449–453.
14. Rojo MG, García GB, Mateos CP, García JG, Vicente MC. Critical comparison of 31 commercially available digital slide systems in pathology. Int J Surg Pathol. 2006; 14:285–305.

15. Farahani N, Parwani AV, Pantanowitz L. Whole slide imaging in pathology: advantages, limitations, and emerging perspectives. Pathol Lab Med Int. 2015; 7:23–33.
16. Baidoshvili A. Making the Move to 100 Percent Digital. Pathologist. 2015; 502–503.
17. Randell R, Ruddle RA, Mello-Thoms C, Thomas RG, Quirke P, Treanor D. Virtual reality microscope versus conventional microscope regarding time to diagnosis: An experimental study. Histopathology. 2013; 62:351–358.

18. Park S, Pantanowitz L, Parwani AV. Digital imaging in pathology. Clin Lab Med. 2012; 32:557–584.

19. Bhattacharyya S. A brief survey of color image preprocessing and segmentation techniques. J Pattern Recognit Res. 2011; 6:120–129.

20. Khan AM, Rajpoot N, Treanor D, Magee D. A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution. IEEE Trans Biomed Eng. 2014; 61:1729–1738.

21. He L, Long LR, Antani S, Thoma GR. Histology image analysis for carcinoma detection and grading. Comput Methods Programs Biomed. 2012; 107:538–556.

22. Chen JM, Li Y, Xu J, Gong L, Wang LW, Liu WL, Liu J. Computer-aided prognosis on breast cancer with hematoxylin and eosin histopathology images: A review. Tumour Biol. 2017; 39:1010428317694550.

23. Parvin B, Yang Q, Han J, Chang H, Rydberg B, Barcellos-Hoff MH. Iterative voting for inference of structural saliency and characterization of subcellular events. IEEE Trans Image Process. 2007; 16:615–623.

24. Al-Kofahi Y, Lassoued W, Lee W, Roysam B. Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans Biomed Eng. 2010; 57:841–852.

25. Arteta C, Lempitsky V, Noble JA, Zisserman A. Learning to detect cells using non-overlapping extremal regions. Med Image Comput Comput Assist Interv. 2012; 15:348–356.

26. Yan C, Li A, Zhang B, Ding W, Luo Q, Gong H. Automated and accurate detection of soma location and surface morphology in large-scale 3D neuron images. PLoS One. 2013; 8:e62579.

27. Esmaeilsabzali H, Sakaki K, Dechev N, Burke RD, Park EJ. Machine vision-based localization of nucleic and cytoplasmic injection sites on low-contrast adherent cells. Med Biol Eng Comput. 2012; 50:11–21.

28. Jung C. Segmenting clustered nuclei using H-minima transform-based marker extraction and contour parameterization. IEEE Trans Biomed Eng. 2010; 57:2600–2604.

29. Li F, Zhou X, Ma J, Wong ST. Multiple nuclei tracking using integer programming for quantitative cancer cell cycle analysis. IEEE Trans Med Imaging. 2010; 29:96–105.

30. Ali S. An integrated region-, boundary-, shapebased active contour for multiple object overlap resolution in histological imagery. IEEE Trans Med Imaging. 2012; 31:1448–1460.

31. Cheng JR. Segmentation of clustered nuclei with shape markers and marking function. IEEE Trans Biomed Eng. 2009; 56:741–748.

32. Basavanhally AN, Ganesan S, Agner S, Monaco JP, Feldman MD, Tomaszewski JE, et al. Computerized image-based detection and grading of lymphocytic infiltration in her2+ breast cancer histopathology. IEEE Trans Biomed Eng. 2010; 57:642–653.

33. Fatakdawala H, Xu J, Basavanhally A, Bhanot G, Ganesan S, Feldman M, et al. Expectation-maximization-driven geodesic active contour with overlap resolution (EMaGACOR): application to lymphocyte segmentation on breast cancer histopathology. IEEE Trans Biomed Eng. 2010; 57:1676–1689.

34. Vink JP, Van Leeuwen MB, Van Deurzen CH, De Haan G. Efficient nucleus detector in histopathology images. J Microsc. 2013; 249:124–135.

35. Xu J, Xiang L, Liu Q, Gilmore H, Wu J, Tang J, Madabhushi A. Stacked sparse autoencoder (SSAE) for nuclei detection on breast cancer histopathology images. IEEE Trans Med Imaging. 2016; 35:119–130.

36. Hoque A, Lippman SM, Boiko IV, Atkinson EN, Sneige N, Sahin A, et al. Quantitative nuclear morphometry by image analysis for prediction of recurrence of ductal carcinoma in situ of the breast. Cancer Epidemiol Biomarkers Prev. 2001; 10:249–259.
37. Qi X, Xing F, Foran DJ, Yang L. Robust segmentation of overlapping cells in histopathology specimens using parallel seed detection and repulsive level set. IEEE Trans Biomed Eng. 2012; 59:754–765.

38. Latson L, Sebek B, Powell KA. Automated cell nuclear segmentation in color images of hematoxylin and eosin-stained breast biopsy. Anal Quant Cytol Histol. 2003; 25:321–331.
39. Veta M, Kornegoor R, Huisman A, Verschuur-Maes AH, Viergever MA, Pluim JP, van Diest PJ. Prognostic value of automatically extracted nuclear morphometric features in whole slide images of male breast cancer. Mod Pathol. 2012; 25:1559–1565.

40. Mouelhi A, Sayadi M, Fnaiech F. A supervised segmentation scheme based on multilayer neural network and color active contour model for breast cancer nuclei detection. In : 2013 international conference on electrical engineering and software applications (ICEESA); 21–23 March; Hammamet, Tunisia. New York: IEEE;2013. 53:p. 1–6.
41. Naik S, Doyle S, Agner S, Madabhushi A, Feldman M, Tomaszewski J. Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology. In : 5th IEEE international symposium on biomedical imaging: from nano to macro; 14–17 May; Paris, France. New York: IEEE;2008. p. 284–287.
42. Basavanhally A, Ganesan S, Feldman M, Shih N, Mies C, Tomaszewski J, et al. Multifield-of-view framework for distinguishing tumor grade in ER+ breast cancer from entire histopathology slides. IEEE Trans Biomed Eng. 2013; 60(8):2089–2099.

43. Mendelsohn ML, Kolman WA, Perry B, Prewitt JM. Morphological analysis of cells and chromosomes by digital computer. Methods Inf Med. 1965; 4:163–167.

44. Wilbur DC, Prey MU, Miller WM, Pawlick GF, Colgan TJ. The AutoPap system for primary screening in cervical cytology. Comparing the results of a prospective, intended-use study with routine manual practice. Acta Cytol. 1998; 42:214–220.
45. Dawson AE. Can We Change the Way We Screen?: The ThinPrep Imaging System. Cancer. 2004; 102:340–344.

46. Mungle T, Tewary S, DAS DK, Arun I, Basak B, Agarwal S, et al. MRF-ANN: a machine learning approach for automated ER scoring of breast cancer immunohistochemical images. J Microsc. 2017; 267:117–129.

47. Tuominen VJ, Ruotoistenmäki S, Viitanen A, Jumppanen M, Isola J. ImmunoRatio: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res. 2010; 12:R56.

48. Albarqouni S, Baur C, Achilles F, Belagiannis V, Demirci S, Navab N. AggNet: Deep Learning From Crowds for Mitosis Detection in Breast Cancer Histology Images. IEEE Trans Med Imaging. 2016; 35:1313–1321.

49. Cireşan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis Detection in Breast Cancer Histology Images with Deep Neural Networks. Med Image Comput Comput Assist Interv. 2013; 16:411–418.

50. Veta M, van Diest PJ, Willems SM, Wang H, Madabhushi A, Cruz-Roa A, et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med Image Anal. 2015; 20:237–248.

51. Wang H, Cruz-Roa A, Basavanhally A, Gilmore H, Shih N, Feldman M, et al. Mitosis detection in breast cancer pathology images by combining handcrafted and convolutional neural network features. J Med Imaging (Bellingham). 2014; 1:034003.

52. Malon CD, Cosatto E. Classification of mitotic figures with convolutional neural networks and seeded blob features. J Pathol Inform. 2013; 4:9.

53. Ali HR, Dariush A, Provenzano E, Bardwell H, Abraham JE, Iddawela M, et al. Computational pathology of pre-treatment biopsies identifies lymphocyte density as a predictor of response to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res. 2016; 18:21.

54. Turkki R, Linder N, Kovanen PE, Pellinen T, Lundin J. Antibody-supervised deep learning for quantification of tumor-infiltrating immune cells in hematoxylin and eosin stained breast cancer samples. J Pathol Inform. 2016; 7:38.

55. Yuan Y. Modelling the spatial heterogeneity and molecular correlates of lymphocytic infiltration in triple-negative breast cancer. J R Soc Interface. 2015; 12:20141153.

56. Roux L, Racoceanu D, Loménie N, Kulikova M, Irshad H, Klossa J, et al. Mitosis detection in breast cancer histological images an ICPR 2012 contest. J Pathol Inform. 2013; 4:8.

57. Giusti A, Cacciay C, Ciresan DC, Schmidhuber J, Gambardella LM. A comparison of algorithms and humans for mitosis detection. In : 2014 IEEE 11th international symposium on biomed imaging (ISBI); 29 April–2 May; Beijing, China. New York: IEEE;2014. p. 1360–1363.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download