Abstract
Transcriptional regulation of a gene is not always correlated with genetic information inherited from parents because the transcription of specific genes is often governed by the modification of chromatin structure. The study of transcriptional regulation by modifying chromatin structure is well-known as "epigenetics". Several methods involved in the modification of chromatin structure have been developed in the mammalian species during evolution. Among those methods, methylations of specific DNA region or histone are often used to control specific gene transcription. Therefore, understanding the activity of proteins involved in DNA or histone methylation is an initial step to control the transcriptional activity of a specific gene. Polycomb group (PcG) proteins were known to be repressors of transcription of a specific gene by creating and maintaining methylation or ubiquitination of the specific region of histone. Dependent on the target histone, the activity of PcG proteins effects on the development of specific lineage cells or the activity of specific cell types. In this review, the function, expression and activity of PcG proteins related with the development or activation of T cells are discussed.
Figures and Tables
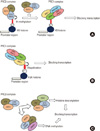 | Fig. 1Transcriptional inhibition by polycomb proteins [10-17]. (A) Trimethylation of K27 residue on H3 histone by polycomb proteins blocks the transcription of target gene, (B) Ubiquitination of K119 residue on H2A histone by polycomb proteins blocks the transcription of target gene, (C) Either histone deacetylation or DNA methylation by polycomb proteins blocks the transcription of target gene. |
References
1. Jaenisch R, Bird A. Eigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003. 33:245–254.

2. Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010. 35:323–332.

3. Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007. 128:735–745.

4. Moazed D, O'Farrell PH. Maintenance of the engrailed expression pattern by Polycomb group genes in Drosophila. Development. 1992. 116:805–810.

5. Kennison JA. The polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet. 1995. 29:289–303.

6. Gould A. Functions of mammalian polycomb group and trithorax group related genes. Curr Opin Genet Dev. 1997. 7:488–494.

7. Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002. 298:1039–1043.

8. Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004. 431:873–878.

9. van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group EED involved histone deaceylation. Nat Genet. 1999. 23:474–478.

10. Hernández-Muñoz I, Taghavi P, Kuijl C, Neefjes J, van Lohuizen M. Association of BMI-1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1. Mol Cell Biol. 2005. 25:11047–11058.

11. Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006. 6:846–856.

12. Nakayama T, Yamashita M. Critical role of the polycomb and trithorax complexes in the maintenance of CD4 T cell memory. Semin Immunol. 2009. 21:78–83.

13. Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002. 16:2893–2905.

14. Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004. 14:155–164.

15. Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003. 17:1870–1881.

16. de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004. 7:663–676.

17. Shilatifard A. Chromatin modifications by methylation and uqiuitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006. 75:243–269.

18. van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994. 8:757–769.

19. Lessard J, Baban S, Sauvageau G. Stage-specific expression of polycomb group genes in human bone marrow cells. Blood. 1998. 91:1216–1224.

20. Haupt Y, Bath ML, Harris AW, Adams JM. Bmi-1 transgene induces lymphomas and collaborates with myc in tumorigenesis. Bmi-1 transgene induces lymphomas and collaborates with myc in tumorigenesis. Oncogene. 1993. 8:3161–3164.
21. Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003. 423:302–305.

22. Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999. 397:164–168.

23. Raaphorst FM, Otte AP, van Kemenade FJ, Blokzijl T, Fieret E, Hamer KM, Satijn DP, et al. Distinct BMI-1 and EZH2 expression patterns in thymocytes and mature T cells suggest a role for Polycomb genes in human T cell differentiation. J Immunol. 2001. 166:5925–5934.

24. Miyazaki M, Miyazaki K, Itoi M, Katoh Y, Guo Y, Kanno R, et al. Thymocyte proliferation induced by pre-T cell receptor signaling is maintained through polycomb gene product Bmi-1-mediated Cdkn2a repression. Immunity. 2008. 28:231–245.

25. Hosokawa H, Kimura MY, Shinnakasu R, Suzuki A, Miki T, Koseki H, et al. Regulation of Th2 cell development by Polycomb group gene bmi-1 through the stabilization of GATA3. J Immunol. 2006. 177:7656–7664.

26. Yamashita M, Kuwahara M, Suzuki A, Hirahara K, Shinnaksu R, Hosokawa H, et al. Bmi1 regulates memory CD4 T cell survival via repression of the Noxa gene. J Exp Med. 2008. 205:1109–1120.

27. Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000. 288:1053–1058.

28. Guo Y, Miyazaki M, Itoi M, Satoh R, Iwama A, Amagai T, et al. Polycomb group gene Bmi1 plays a role in the growth of thymic epithelial cells. Eur J Immunol. 2011. 41:1098–1107.

29. Akasaka T, Tsuji K, Kawahira H, Kanno M, Harigaya K, Hu L, et al. The role of mel-18, a mammalian Polycomb group gene, during IL-7-dependent proliferation of lymphocyte precursors. Immunity. 1997. 7:135–146.

30. Foxwell BM, Beadling C, Guschin D, Kerr I, Cantrell D. Interleukin-7 can induce the activation of Jak 1, Jak 3 and STAT 5 proteins in murine T cells. Eur J Immunol. 1995. 25:3041–3046.

31. Tetsu O, Ishihara H, Kanno R, Kamiyasu M, Inoue H, Tokuhisa T, Taniguchi M, Kanno M. mel-18 negatively regulates cell cycle progression upon B cell antigen receptor stimulation through a cascade leading to c-myc/cdc25. Immunity. 1998. 9:439–448.

32. Kimura M, Koseki Y, Yamashita M, Watanabe N, Shimizu C, Katsumoto T, et al. Regulation of Th2 cell differentiation by mel-18, a mammalian polycomb group gene. Immunity. 2001. 15:275–287.

33. Chou RH, Yu YL, Hung MC. The roles of EZH2 in cell lineage commitment. Am J Transl Res. 2011. 3:243–250.
34. Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007. 39:232–236.

35. O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001. 21:4330–4336.
36. Koyanagi M, Baguet A, Martens J, Margueron R, Jenuwein T, Bix M. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J Biol Chem. 2005. 280:31470–31477.

37. Li G, Yu M, Weyand CM, Goronzy JJ. Epigenetic regulation of killer immunoglobulin-like receptor expression in T cells. Blood. 2009. 114:3422–3430.

38. Ogawa M, Hiraoka Y, Aiso S. The Polycomb-group protein ENX-2 interacts with ZAP-70. Immunol Lett. 2003. 86:57–61.

39. Herrera-Merchan A, Arranz L, Ligos JM, de Molina A, Dominguez O, Gonzalez S. Ectopic expression of the histone methyltransferase Ezh2 in haematopoietic stem cells causes myeloproliferative disease. Nat Commun. 2012. 3:623.

40. Katoh-Fukui Y, Owaki A, Toyama Y, Kusaka M, Shinohara Y, Maekawa M, et al. Mouse Polycomb M33 is required for splenic vascular and adrenal gland formation through regulating Ad4BP/SF1 expression. Blood. 2005. 106:1612–1620.

41. Suzuki A, Iwamura C, Shinoda K, Tumes DJ, Kimura MY, Hosokawa H, et al. Polycomb group gene product Ring1B regulates Th2-driven airway inflammation through the inhibition of Bim-mediated apoptosis of effector Th2 cells in the lung. J Immunol. 2010. 184:4510–4520.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download