Abstract
Recent progress of genetics has dramatically improved pharmacogenetics for human diseases. Several pharmacogenetic assays such as TPMT/azathioprine and CYP2C9/VKORC1/warfarin have been introduced in clinical practice at the present time. Rheumatoid arthritis (RA) is a chronic systemic autoimmune disease that leads to irreversible joint damage and disability if it is not adequately treated. Although the introduction of anti-TNF therapy has improved the outcome of therapy in RA, a substantial proportion of patients (approximately 30-40%) fail to respond to the drugs. Recently, pharmacogenetic studies have widely been performed to search for genetic and mRNA expression biomarkers to predict the response of anti-TNF therapy in RA. Other potential serum biomarkers of response have also been explored including cytokines and autoantiboides. None has yet been validated for biomarkers that predict the response of biologic drugs in clinical rheumatology practice. However, future medicine using pharmacogenetic applications in RA might make personalized therapy possible.
Figures and Tables
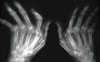 | Fig. 1Simple x-rays of hands of patients with rheumatoid arthritis shows the ulnar deviation of both hands, joint space narrowing of wrist and metacarpophalangeal joints, and periarticular osteopenia. |
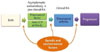 | Fig. 2Genetic and environmental factors influence on both stages of rheumatoid arthritis (RA) development, asymptomatic autoantibody positive preclinical RA and clinical RA. |
Table 2
Recommended Daily Warfarin doses (mg/day) to Achieve a Therapeutic INR Based on CYP2C9 and VKORC1 Genotype using the Warfarin Product Insert Approved by the United States Food and Drug Administration. Reproduced from Updated Warfarin (Coumadin®) Product label [6]

Ref. 6 with permission from Nature Publishing Group
Table 4
Clinical Markers to Predict the Response of anti-TNF Therapy

DMARD, disease modifying anti-rheumatic drug; DAS28, Disease Activity Score 28; EULAR, The European League Against Rheumatism; ACR, American College of Rheumatology; HAQ, Health assessment questionnaire
Ref. 15 with permission from BMJ Publishing Group
Table 6
Pharmacogenetic study for anti-TNF therapy in rheumatoid arthritis (candidate gene approach) (*E-Etanercept, I-Infliximab, A-Adalimumab) [19]

TNF, Tumor necrosis factor; ACR, American College of Rheumatology; DAS28, Disease Activity Score 28; EULAR, The European League Against Rheumatism
Ref. 19 with permission from Future Medicine
References
1. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001. 409:860–921.
2. International HapMap Consortium. A haplotype map of the human genome. Nature. 2005. 437:1299–1320.
3. 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010. 467:1061–1073.
4. Pirmohamed M, James S, Meakin S, Green C, Scott AK, Walley TJ, et al. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18 820 patients. BMJ. 2004. 329:15–19.

5. Wright C, Burton H, Hall A, Moorthie S, Pokorska-Bocci A, Sanderson S, et al. Next steps in the sequence. The implications of whole genome sequencing for health in the UK. 2011. Cambridge: Foundation for Genomics and Population Health(PHG);ISBN 978-1-907198-08-3.
6. Johnson JA, Gong L, Whirl-Carrillo M, Gage BF, Scott SA, Stein CM, et al. Clinical Pharmacogenetics Implementation Consor tium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther. 2011. 90:625–629.

7. Relling MV, Gardner EE, Sandborn WJ, Schmiegelow K, Pui CH, Yee SW, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin Pharmacol Ther. 2011. 89:387–391.

8. Martin MA, Klein TE, Dong BJ, Pirmohamed M, Haas DW, Kroetz DL. Clinical Pharmacogenetics Implementation Consortium Guidelines for HLA-B Genotype and Abacavir Dosing. Clin Pharmacol Ther. 2012. 91:734–738.

9. Daly AK, Donaldson PT, Bhatnagar P, Shen Y, Pe'er I, Floratos A, et al. HLA-B[ast]5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat Genet. 2009. 41:816–819.

10. Callahan R, Hurvitz S. Human epidermal growth factor receptor-2-positive breast cancer: Current management of early, advanced, and recurrent disease. Curr Opin Obstet Gynecol. 2011. 23:37–43.

11. Wang L, McLeod HL, Weinshilboum RM. Genomics and Drug Response. N Engl J Med. 2011. 364:1144–1153.

12. Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO 3rd, et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann Rheum Dis. 2010. 69:1580–1588.

13. Visser K, Goekoop-Ruiterman YP, de Vries-Bouwstra JK, Ronday HK, Seys PE, Kerstens PJ, et al. A matrix risk model for the prediction of rapid radiographic progression in patients with rheumatoid arthritis receiving different dynamic treatment strategies: post hoc analyses from the BeSt study. Ann Rheum Dis. 2010. 69:1333–1337.

14. Smolen JS, Landewé R, Breedveld FC, Dougados M, Emery P, Gaujoux-Viala C, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs. Ann Rheum Dis. 2010. 69:964–975.
15. Emery P, Dörner T. Optimising treatment in rheumatoid arthritis: a review of potential biological markers of response. Ann Rheum Dis. 2011. 70:2063–2070.

16. Potter C, Hyrich KL, Tracey A, Lunt M, Plant D, Symmons DP, et al. Association of rheumatoid factor and anti-cyclic citrullinated peptide positivity, but not carriage of shared epitope or PTPN22 susceptibility variants, with anti-tumour necrosis factor response in rheumatoid arthritis. Ann Rheum Dis. 2009. 68:69–74.

17. Hyrich KL, Lunt M, Watson KD, Symmons DP, Silman AJ. British Society for Rheumatology Biologics Register. Outcomes after switching from one anti-tumor necrosis factor alpha agent to a second anti-tumor necrosis factor alpha agent in patients with rheumatoid arthritis: results from a large UK national cohort study. Arthritis Rheum. 2007. 56:13–20.

18. Takeuchi T, Miyasaka N, Tatsuki Y, Yano T, Yoshinari T, Abe T, et al. Baseline tumour necrosis factor alpha levels predict the necessity for dose escalation of infliximab therapy in patients with rheumatoid arthritis. Ann Rheum Dis. 2011. 70:1208–1215.

19. Prajapati R, Plant D, Barton A. Genetic and genomic predictors of anti-TNF response. Pharmacogenomics. 2011. 12:1571–1585.

20. Plant D, Bowes J, Potter C, Hyrich KL, Morgan AW, Wilson AG, et al. Genome-wide association study of genetic predictors of anti-tumor necrosis factor treatment efficacy in rheumatoid arthritis identifies associations with polymorphisms at seven loci. Arthritis Rheum. 2011. 63:645–653.

21. Julià A, Erra A, Palacio C, Tomas C, Sans X, Barceló P, et al. An eight-gene blood expression profile predicts the response to infliximab in rheumatoid arthritis. PLoS One. 2009. 4:e7556.





 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download