Abstract
The H9N2 subtype low pathogenic avian influenza is one of the most prevalent avian diseases worldwide, and was first documented in 1996 in Korea. This disease caused serious economic loss in Korea's poultry industry.
In order to develop an oil-based inactivated vaccine, a virus that had been isolated in 2001 (A/chicken/Korea/01310/2001) was selected based on its pathogenic, antigenic, and genetic properties. However, in animal experiments, the efficacy of the vaccine was found to be very low without concentration of the antigen (27 to 210 hemagglutinin unit). In order to overcome the low productivity, we passaged the vaccine candidate virus to chicken eggs. After the 20th passage, the virus was approximately ten times more productive compared with the parent virus. For the most part, the passaged virus maintained the hemagglutinin cleavage site amino acid motif (PATSGR/GLF) and had only three amino acid changes (T133N, V216G, E439D, H3 numbering) in the hemagglutinin molecule, as well as 18 amino acid deletions (55-72) and one amino acid change (E54D) in the NA stalk region. The amino acid changes did not significantly affect the antigenicity of the vaccine virus when tested by hemagglutination inhibition assay. Though not complete, the vaccine produced after the 20th passage of the virus (01310 CE20) showed good protection against a homologous and recent Korean isolate (A/chicken/Korea/Q30/2004) in specific pathogen- free chickens.
The vaccine developed in this study would be helpful for controlling the H9N2 LPAI in Korea.
Avian influenza virus (AIV) is an enveloped virus that belongs to the Orthomyxoviridae family and has an eight segmented, single stranded, negative sense RNA genome. Among the proteins encoded by the genome, there are two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA). AIV is classified into subtypes according to the combination of 16 HA and 9 NA molecules [10,26].
Among the many subtypes, the H9N2 AIV is thought to have originated from shorebirds and gulls [30], and rapidly spread to become one of the most prevalent diseases in domestic poultry worldwide. It has also caused serious economic loss in the poultry industry [5,16,17]. In Korea, the first H9N2 low pathogenic avian influenza (LPAI) outbreak occurred in 1996 [15]. Since 2000, and it has become endemic (especially in layer farms) [13,16]. Many studies have demonstrated that there are several distinct H9N2 AIV lineages, and indicated the Korean H9N2 viruses formed a unique antigenic and phylogenetic cluster [13,15-18].
Although immunization with this vaccine is not complete, it is one of the most promising control measures for the H9N2 LPAI to date. Some countries have used vaccines for H9N2 LPAI [5,17,22,29]; however, vast antigenic variations exist even within the same subtype, and it is very difficult to select a vaccine strain that is effective on the virus in current circulation. In addition, some isolates do not grow to a high enough titer in the embryonated chicken eggs (ECEs) to achieve efficient vaccine productivity [31].
Korean animal health authorities took stamping-out and compensation control policies with regards to H9N2 LPAI when it occurred between 1996 and 1999. At that time, vaccines for subtypes of AIV, including H9N2 LPAI, were prohibited in Korea because they interfered in the discrimination of naturally infected birds from vaccinated birds. However, H9N2 LPAI became endemic, and the policy was not reliable enough to cover each outbreak. According to the 2004 Avian influenza standard operating procedures, Korean animal health authorities permitted the use of the vaccine for LPAI (especially the H9N2 subtype), and the Committee on the National AI Vaccine Campaign determined that using a single vaccine strain was the most effective strategy with which to simplify the H9N2 AI situation in Korea [7,20].
In this study, we present the characterization of the Korean H9N2 LPAI vaccine strain, and evaluated the efficacy of the pilot vaccine in specific pathogen-free (SPF) chickens.
All of the viruses were isolated by the National Veterinary Research and Quarantine Service using routine diagnostic practices. The infectious tissue homogenates were inoculated in the allantoic cavity of 9-11 day old SPF ECEs (Lohmann Valo SPF Cuxhaven, Germany) according to standard procedures [27]. The first H9N2 isolate in Korea, A/Chicken/Korea/MS96/1996 (MS96) [15] was used, and A/Chicken/Korea/99029/1999 (99029) and A/Chicken /Korea/01310/2001 (01310) were used as representative isolates of the 1999 and 2001 strain, respectively. The 2001 strain was eventually chosen as the vaccine candidate. In order to test the antigenicity and the efficacy of the vaccine, a recent isolate, A/Chicken/Korea/Q30/2004 (04Q30), was used [16].
In order to determine the pathogenicity of the selected viruses in SPF and commercial broiler chickens, MS96, 99029, and 01310 were inoculated via the intra-tracheal route in eight 7-week-old SPF chickens (106.5EID50/0.1 ml, 105.6 EID50/0.1 ml, and 107.1 EID50/0.1 ml, respectively) and fifteen 12-week-old commercial broiler chickens (105.2 EID50/0.1 ml, 105.7 EID50/0.1 ml, and 105.0 EID50/0.1 ml, respectively), which were confirmed to be free of antibodies against H9N2 AIV. In the experiment with SPF chickens, tracheal and cloacal swab samples were taken at 3, 5, 7, and 9 days post-inoculation (dpi). The swab samples were suspended in 3 ml of gentamicin-PBS (1% gentamicin in PBS, pH 7.2), and were inoculated with 0.2 ml of samples into three 9-11 day old SPF ECEsvia the allantoic cavity route. The inoculated broiler chickens were reared for 2 weeks, and the mortality was recorded.
After being propagated in ECEs, the viruses (MS96, 99029, and 01310) were inactivated by incubating them with 0.1% formalin at 20℃ for 10 h. The inactivation was confirmed by injecting formalin-treated virus into the allantoic cavities of 10-day-old ECEs, two times serially. Virus inactivation was determined by hemagglutination negativity, using 1% chicken red blood cells. The inactivated virus was emulsified with oil adjuvant (Montanide ISA 70 SEPPIC, France) at a ratio of 3:7, and wasinjected into eight 6-week-old SPF chickens. The antisera were obtained at 3 weeks after injection. In order to determine the antigenic relationship between the selected viruses, we performed a cross hemagglutination inhibition (HI) test with each of the antisera and virus antigens (four HA unit), and the r-value was subsequently calculated [2].
Viral RNA was extracted from infectious allantoic fluid using the Viral Gene-Spin Viral DNA/RNA extraction kit (iNtRON Biotechnology, Korea) and amplified using gene-specific primer sets by RT-PCR with a Qiagen one-step RT-PCR kit (Qiagen, USA) according to the method described by Hoffmann et al. [12]. The amplified product was excised from agarose gel and eluted using the GENECLEAN SPIN kit (Qbiogene, USA). The nucleotide sequences were analyzed by direct sequencing of the PCR products using ABI PRISM BigDye Terminator Cycle Sequencing Kits (Applied Biosystems, USA). The HA and NA gene nucleotide sequences of 01310 CE3 have been deposited in GenBank under accession number EU253561 and EU253562, respectively.
The nucleotide and deduced amino acid sequences of the HA and NA molecules were aligned by the Clustal W method with the MegAlign software (Lasergene 7.0; DNASTAR, USA). The similarity of the HA and NA amino acid sequences were compared with recently studied Korean H9N2 AIVs [16].
The selected vaccine candidate (01310 CE3, 27 HA unit) virus based on the above pathogenic, serologic, and molecular data (refer to the Results section) was used to prepare the vaccine according to the previously mentioned procedures. The preparation of the high HA content vaccine (210 HA unit) involved the concentration of the virus by centrifugation (18,000 rpm, 4 h; Beckman, USA). The vaccines were injected into ten 6-week-old SPF chickens via the intramuscular (IM) route. The serum samples were taken three weeks post-vaccination (wpv), and were followed by the performance of the HI test. The HI test results were analyzed using Student's t-test. Statistical significance was set a priori at α = 0.05. The chickens were challenged with 01310 CE3 virus (106.0 EID50/0.1 ml) at 3 wpv via the oral route. Next, oropharyngeal and cloacal swab samples were taken at 5 dpi for virus isolation. The Fisher's exact test was performed to compare the virus isolation rate (α= 0.05).
In order to recover the highly growing vaccine virus in ECEs, the vaccine candidate virus was serially passaged through 9- to 11-day-old SPF ECEs and selected with high HA titer using chicken red blood cells. The passage number in ECEs was indicated as CE X (X stands for passage number) after the virus name.
In order to test the changes in the pathogenicity of the passaged vaccine viruses in chicken eggs, the viruses (CE5, CE10, CE15, CE20, and CE40) were diluted in PBS and inoculated in the allantoic cavities of 10-day-old SPF ECEs. In addition, the mortality was checked after 48 h of incubation.
For the pathogenicity test of the selected vaccine virus in chickens, eight 6-week-old SPF chickens were intravenously inoculated with 0.2 ml of 1/10 diluted virus, and we noted the mortality for 10 days according to standard procedures [31].
The vaccines of the antigenicity test were prepared with 01310 CE6, CE20, and CE40 viruses. In addition, the HA titers of the viruses were adjusted to 29 HA units. Eight 6-week-old SPF chickens were then vaccinated at 0.5 ml/chickenvia the IM route. After vaccination, serum samples were taken at 3 wpv, and the HI assay was then performed with the MS96 CE3, 01310 CE20, and 04Q30 CE3 viruses. A one-way ANOVA test was subsequently performed.
In order to determine the optimum growth conditions of the vaccine strain in ECEs, the virus was serially diluted 10-fold with 1% gentamicin-PBS (10-3-10-7), and was inoculated into 60 SPF ECEs. At 8, 16, 24, 32, 40, and 48 hours after incubation, 10 eggs were harvested during each dilution and the HA titer was determined with 1% chicken red blood cells [27].
The vaccine was prepared with the 01310 CE20 (210 HA unit) strain as per the procedures mentioned in the Materials and Methods section, and was injected at a concentration of 0.5 ml/chicken via the intramuscular route (thigh muscle) into each of eight 6-week-old SPF chickens. Three weeks after vaccination, the parent virus (01310 CE3, 105.5 EID50/0.1 ml) and recent Korean isolate (04Q30 CE3, 105.5 EID50/0.1 ml) were challenged intranasally. To isolate the challenged virus, oropharyngeal and cloacal swab samples were taken at 1, 3, and 5 dpi from each chicken and suspended in 3 ml of gentamicin-PBS. The samples were subsequently inoculated into three 9- to 11-day-old SPF ECEs respectively, followed by assessment of the virus growth from the allantoic fluids. At 5 dpi, the chickens were sacrificed and the various tissues were taken with different scissors in order to prevent contamination. Next, the 10% (w/v) tissue homogenates were tested for viral growth in three 9- to 11-day-old SPF ECEs. In order to titrate the virus from various tissues, tissue homogenates were pooled in a group of equal volume, and were then titrated using SPF ECEs.
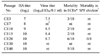
In order to select the vaccine candidate, we chose the MS96, 99029, and 01310 viruses as the representative viruses of the year. In addition, we compared the virus replication potency and pathogenicity for SPF and commercial broiler chickens. For the SPF chickens, all of the viruses had a zero mortality rate; however, the viruses were isolated at 3 to 5 dpi from oropharyngeal swab samples, and 3 to 7 dpi from cloacal swab samples. At 5 dpi, the MS96, 99029, and 01310 viruses were isolated from 5/8, 7/8, and 6/8 oropharyngeal swab samples and 1/8, 3/8, and 5/8 cloacal swab samples, respectively. Furthermore, in commercial broiler chickens, the 01310 strain showed a 26.7% mortality rate, whereas a 6.7% mortality rate was recorded for each of the other two viruses (Table 1).
To elucidate the antigenic relationship between the selected Korean isolates, the r-value was calculated with a cross HI titer between the viruses. The values ranged from 0.61 to 1.00, with no significant differences between the viruses (Table 2).
Based on our pathogenic and serologicdata, we chose the 01310 virus as a vaccine candidate. Moreover, when we compared the HA and NA amino acid sequences deduced by nucleotide sequences, the 01310 CE3 virus was found to have 95.5-98.6% and 94.6-98.5% similarity with the recent Korean H9N2 AIVs isolated from 2002 to 2004, respectively (data not shown).
In order to elucidate the efficacy of the vaccine candidate virus, ten SPF chickens were immunized with different antigen contents. Chickens immunized with a high antigen vaccine (210 HA unit) showed a similar but statistically significant (p < 0.05) antibody titer compared to the low antigen content (27 HA unit) vaccine (Table 3). In addition, in the low antigen content vaccine group, the challenged viruses were recovered from 7/10 and 9/10 chickens from oropharyngeal and cloacal swab samples at 5 dpi, respectively. It was believed that the vaccine was not effective to protect against viral shedding. However, in high antigen content groups, the challenged virus was isolated from 1/10 or 3/10 oropharyngeal/cloacal swab samples, respectively. Although neither of the vaccines were able to completely protect against viral shedding, the high antigen content vaccine was more effective than the low antigen content vaccine upon comparison of the virus recovery rate in the oropharyngeal/cloacal swab samples (p = 0.02, respectively) (Table 3).
The vaccine candidate virus (01310 CE3) grew in ECEs at around 27 HA units and about 107.0 EID50/0.1 ml, as did most of the field isolates (personal observation, data not presented), and we were not able to determine the expected vaccine efficacy with unconcentrated antigen (Table 3). Therefore, we attempted to get highly growing phenotyped viruses through egg passage. When the 01310 strain was passaged in SPF ECEs as a vaccine candidate, the virus titer increased with each ECE passage (Table 4). After the 15th passage in ECEs, the virus titer showed 210 HA units stably; this was approximately 10 times greater than that of the parent virus. Moreover, the 20th passaged virus showed the highest titer, 108.7 EID50/0.1 ml (Table 4). As shown in Fig. 1, the 01310 CE20 showed the highest HA titer when inoculated with 104.7 EID50/0.1 ml and incubated for 32 hours. The mean HA titer at that point was 9.7 ± 0.5 (log2).
However, as the 01310 virus was passaged in SPF ECEs, the mortality of the chicken eggs also increased. During 48 h of incubation, sixof ten eggs infected with the 01310 CE20 virus were dead, whereas, two to three of ten eggs were dead due to the CE15 virus or other less passaged viruses. In spite of increased mortality in chicken eggs, the CE20 virus showed no mortality for the intravenous challenge experiment with SPF chickens, and was thought to maintain the LPAI characteristics according to the OIE criteria [31] (Table 4).
Upon comparison of HA amino acid sequences of the 01310 CE3, CE20, and CE40 to HA cleavage (PATSGR/GLF), receptor binding (183H, 190E, 226Q, 228G), and potential glycosylation sites (158N), we found no changes (even in the CE40 passaged virus). HA has only three amino acid changes in the CE20 virus (T133N, V216G in HA1 region, and E439D in HA2 region), and an additional change occurred (L531F) in the CE40 virus (H3 numbering). In the NA molecule, the CE20 and CE40 viruses have an 18 amino acid deletion in the stalk region (55-72); both of these viruses also contain a change in one amino acid (E54D) (data not shown).
As shown in Table 5, the HI titers (log2) of the 01310 CE20 antisera with MS96 CE3, 01310 CE20, and the recent isolate, 04Q30 CE3, were 8.8 ± 0.5, 8.3 ± 0.5, and 9.1 ± 1.0, respectively. Moreover, the HI titers of antisera of 01310 CE6, CE20, and CE40 with the homologous Ag (01310 CE20) were 7.8 ± 0.2, 8.3 ± 0.5, and 8.4 ± 0.6, respectively. One-way ANOVA comparison of the HI titers revealed no significant difference.
For the vaccine efficacy test, eight 6-week-old chickens were vaccinated with 01310 CE20 and challenged after 3 weeks with homologous parent vaccine virus (01310 CE3) or the recent Korean isolate (04Q30 CE3). Moreover, swab samples were taken at 1, 3, and 5 dpi, and were tested for virus isolation. The highest isolation number was obtained for 5 dpi in both of the unvaccinated groups. The oropharyngeal and cloacal swab samples taken at 5 dpi showed isolation of 7/8 and 6/8 for the 01310 CE3 group and 8/8 and 5/8 for the 04Q30 CE3 group, respectively. On the other hand, the viruses for the vaccinated group, which were isolated only from the 04Q30 challenge group, showed isolation of 2/8 oropharyngeal swab samples taken at 1 and 3 dpi (Table 6).
In the tissue samples taken at 5 dpi, the challenged viruses were not isolated from the vaccinated group, whereas for the unvaccinated group, the 01310 CE3 virus was isolated from trachea (5/8, 101.4 EID50/0.1ml), spleen (1/8, 0), and cecal tonsil (4/8, 105.4), but not from the brain, lung, or kidney. In addition, the 04Q30 CE3 virus was isolated from the trachea (8/8, 104.4), lung (6/8, 102.0), spleen (2/8, 0), kidney (2/8, 101.6), and cecal tonsil (5/8, 106.0), but not from the brain. In both of the unvaccinated groups, the cecal tonsil showed the highest viral titer (Table 6).
Since the first H9N2 LPAI outbreak in 1996, numerous cases have occurred in Korea, and the H9N2 subtype LPAI has become one of the greatest problems of the country's poultry industry (especially for breeders and commercial layers) [13,16].
It is worth noting that avian influenza viruses in their natural hosts, which include wild waterfowl, gulls, and shorebirds, have shown a high rate of genetic conservation. Transmission to other species such as poultry may cause significant amounts of genetic and antigenic changes [12,16-18,25,30]. For a vaccine to be effective, it is necessary that the strain has genetic and antigenic traits similar to those of the currently circulating field viruses. Therefore, it is very important to choose the most effective vaccine strain to prevent the current circulation of viruses. Some countries such as China [17], Pakistan [22], Iran [29], and Israel [5] have already begun the use of H9N2 LPAI vaccines. Because of the above mentioned reasons, each of the countries used their own vaccine strains to control the H9N2 LPAI. Based on the properties of AIV, we chose a candidate vaccine virus with a relatively high pathogenicity in chickens, as well as antigenic and genetic similarity to recent field isolates. Despite this, the vaccine efficacy test of the selected virus (01310), according to the antigen contents using unconcentrated antigen (27 HA unit), showed unsatisfactory protective results compared with the high antigen content vaccine. As for other vaccines, the antigen contents are the critical component of an effective vaccine. Despite this, the antigen concentration requires an increased vaccine production cost, which may be a problem in terms of AI vaccination at the farm level. In order to overcome the low titer of the vaccine candidate strain, the virus was passaged in chicken eggs. Consequent to the passages of the selected virus, the vaccine strain may have approximately ten times higher titers than the parent virus. Moreover, the rapid growth property and antigenicity of the virus were maintained through the fortieth passage.
In order to determine the optimum inactivation conditions, the virus was incubated in 0.1% formalin at 37℃. However, under these conditions, the HA titer of the virus was decreased by four times, even after 1 h of incubation (data not shown). HA is considered the major antigenic protein, and a decreased HA titer signifies decreased antigenicity. Therefore, the virus was treated at a lower temperature (20℃) and the inactivation conditions were determined. At 20℃, a 6-hour incubation time is enough to inactivate the virus and not show any HA titer changes, even after 12 h of incubation (data not shown).
Although the HA cleavage site (PATSGR/GLF) was well conserved and showed low pathogenic characteristics in the vaccine strain (01310 CE20), the pathogenicity in ECEs was increased. Therefore, we tested the pathogenicity of the vaccine strain in SPF chickens. When the vaccine strain was injected into SPF chickens via the intravenous route according to the standard procedures [31], no mortality was observed in the SPF chickens. Moreover, the vaccine strain has an amino acid sequence consistent withavian virus characteristics at the receptor binding site (Gln226), which confers a high binding affinity to the 2, 3-linked sialic acid (SA) moieties (abundant in avian and horses) rather than the 2, 6-linked SA moieties found in most mammals [19]. Although it is not sufficient, the pathogenicity for chickens and the low binding potential to 2, 6-linked SA secured the safety of the vaccine strain with regard to human infection (especially in vaccine producers).
We observed three amino acid changes (Thr133Asn, Val216Gly, Glu439Asp) compared to the parent virus (01310 CE3) in the HA molecules. The changed amino acids have similar traits (Thr and Asn have a polar uncharged side chain, Val and Gly have a non-polar side chain, Glu and Asp are acidic amino acids) with the parent sequences. However, the Thr133Asn change created an additional potential N-linked glycosylation site located on the right edge of the receptor binding pocket [11]. Many studies have demonstrated that the addition of oligosaccharides to the HA molecule changes the antigenicity of the virus [1,6,24,28]. However, in this study, the additional potential glycosylation site did not significantly affect the antigenicity of the vaccine strain.
A number of studies have shown that a balance between the HA and NA activities is critical for the efficiency of viral growth in host cells [3,4,8,21,23]. In this study, the HA molecule of the vaccine virus (01310 CE20) has a potential glycosylation site on amino acid 158, and represents an additional potential glycosylation site in the vicinity of the receptor binding site (133Asn), in addition to the 18 amino acid deletion in the NA stalk region. It is not clear that such changes exclusively affect the high growth trait of the vaccine virus. Further studies would be needed to clarify the mechanism for the modification of this trait of the vaccine virus as the virus is being passaged in ECEs. In addition, it is believed that the 54 base pair deletion in the NA gene could be used as a vaccine strain marker to identify the vaccine virus.
Although the vaccine candidate virus was isolated in 2001 and may be thought of as outdated, the virus showed high antigenic and genetic similarity to the recent Korean H9N2 LPAIVs. Moreover and most importantly, the vaccine efficacy test with SPF chickens showed that the vaccine candidate sufficiently protected the chickens from the virus [both the homologous (01310 CE3) and recent Korean isolate (04Q30)] (Table 6). The challenge virus was recovered from only two of the eight oropharyngeal swab samples taken at 1 and 2 dpi in the 04Q30 challenged group, respectively, whereas the challenged virus was recovered throughout the experiment and peaked at 5 dpi for the unvaccinated group. Moreover, in subsequent repeated animal experiments, we were able to obtain a reliable and constant virus recovery rate for the cecal tonsil (data not shown). So far, no standards exist for the evaluation of the inactivated AI vaccine efficacy. Based on the animal challenge experiments (the experiments conducted in our lab and the data not presented), the Committee on the National AI Vaccine Campaign determined that the minimum efficacy requirements for inactivated H9N2 LPAI vaccines was a greater than 80% inhibition of the virus recovery rate in cecal tonsils of the vaccinated group, compared to the rate observed in unvaccinated chickens at 5 dpi.
Despite these requirements, AIV is a continuously evolving virus [9,13,16,17,25,30], and as shown in this study, the inactivated LPAI vaccine cannot completely protect against viral infection and shedding into the environment. In addition, immune pressures (such as vaccination) have resulted further complication of the AI situation [14]. Therefore, the Korean government has conditionally issued a vaccine production license according to the following stipulations: 1) The vaccine producers have to record all selling activity and submit records to the Korean animal health authority (NVRQS) as requested; 2) 20 to 30 sentinel birds must be deployed in every vaccinated poultry farm, and the sera from 10% of the vaccinated birds must be tested with those of unvaccinated sentinel birds in order to monitor LPAI infection at least twice a year, and the results must be submitted to the NVRQS. In addition, as a separate complementary measure, active surveillance has been deployed to monitor any changes in the antigenicity of the current circulating H9N2 LPAIV; if changes in antigenicity are detected, the vaccine virus will be changed to a more appropriate virus.
Figures and Tables
 | Fig. 1Growth curves of the vaccine strain (01310 CE20) according to the virus titer (log10EID50/0.1 ml) of the inocula. |
Table 1
Comparison of virus isolation and mortality of recent Korean H9N2 LPAIV

*7-week-old SPF chickens were inoculated via the intra-tracheal route with MS96 (106.5EID50), 99029 (105.6EID50), and 01310 (107.1EID50): virus isolation/total inoculated. †12-week-old AIV antibody-free commercial broiler chickens were inoculated via the intra-tracheal route with MS96 (105.2 EID50), 99029 (105.7EID50), and 01310 (105.0EID50): number of dead/total inoculated (% mortality). op: oropharyngeal, cl: cloacal.
Table 4
Comparison of virus titer and mortality in ECEs and SPF chickens of passaged vaccine candidate
Acknowledgments
This research was supported by the National Veterinary Research and Quarantine Service, Ministry of Agriculture and Forestry of Korea (Project M-AD15-2005-06-01).
References
1. Abe Y, Takashita E, Sugawara K, Matsuzaki Y, Muraki Y, Hongo S. Effect of the addition of oligosaccharides on the biological activities and antigenicity of influenza A/H3N2 virus hemagglutinin. J Virol. 2004. 78:9605–9611.

2. Archetti I, Horsfall FL Jr. Persistent antigenic variation of influenza A viruses after incomplete neutralization in ovo with heterologous immune serum. J Exp Med. 1950. 92:441–462.

3. Asaoka N, Tanaka Y, Sakai T, Fujii Y, Ohuchi R, Ohuchi M. Low growth ability of recent influenza clinical isolates in MDCK cells is due to their low receptor binding affinities. Microbes Infect. 2006. 8:511–519.

4. Baigent SJ, McCauley JW. Glycosylation of haemagglutinin and stalk-length of neuraminidase combine to regulate the growth of avian influenza viruses in tissue culture. Virus Res. 2001. 79:177–185.

5. Banet-Noach C, Perk S, Simanov L, Grebenyuk N, Rozenblut E, Pokamunski S, Pirak M, Tendler Y, Panshin A. H9N2 influenza viruses from Israeli poultry: a five-year outbreak. Avian Dis. 2007. 51:Suppl 1. 290–296.

6. Banks J, Speidel EC, McCauley JW, Alexander DJ. Phylogenetic analysis of H7 hemagglutinin subtype influenza A viruses. Arch Virol. 2000. 145:1047–1058.
7. Capua I. Vaccination for notifiable avian influenza in poultry. Rev Sci Tech. 2007. 26:217–227.
8. Castrucci MR, Kawaoka Y. Biologic importance of neuraminidase stalk length in influenza A virus. J Virol. 1993. 67:759–764.

9. Choi YK, Seo SH, Kim JA, Webby RJ, Webster RG. Avian influenza viruses in Korean live poultry markets and their pathogenic potential. Virology. 2005. 332:529–537.

10. Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol. 2005. 79:2814–2822.

11. Ha Y, Stevens DJ, Skehel JJ, Wiley DC. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc Natl Acad Sci USA. 2001. 98:11181–11186.

12. Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001. 146:2275–2289.

13. Kwon HJ, Cho SH, Kim MC, Ahn YJ, Kim SJ. Molecular epizootiology of recurrent low pathogenic avian influenza by H9N2 subtype virus in Korea. Avian Pathol. 2006. 35:309–315.

14. Lee CW, Senne DA, Suarez DL. Effect of vaccine use in the evolution of Mexican lineage H5N2 avian influenza virus. J Virol. 2004. 78:8372–8381.

15. Lee CW, Song CS, Lee YJ, Mo IP, Garcia M, Suarez DL, Kim SJ. Sequenceanalysis of the hemagglutinin gene of H9N2 Korean avian influenza viruses and assessment of the pathogenic potential of isolate MS96. Avian Dis. 2000. 44:527–535.

16. Lee YJ, Shin JY, Song MS, Lee YM, Choi JG, Lee EK, Jeong OM, Sung HW, Kim JH, Kwon YK, Kwon JH, Kim CJ, Webby RJ, Webster RG, Choi YK. Continuing evolution of H9 influenza viruses in Korean poultry. Virology. 2007. 359:313–323.

17. Li C, Yu K, Tian G, Yu D, Liu L, Jing B, Ping J, Chen H. Evolution of H9N2 influenza viruses from domestic poultry in Mainland China. Virology. 2005. 340:70–83.

18. Liu JH, Okazaki K, Mweene A, Shi WM, Wu QM, Su JL, Zhang GZ, Bai GR, Kida H. Genetic conservation of hemagglutinin gene of H9 influenza virus in chicken population in Mainland China. Virus Genes. 2004. 29:329–334.

19. Matrosovich MN, Krauss S, Webster RG. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology. 2001. 281:156–162.

20. Ministry of Agriculture and Forestry (MAF). Avian Influenza Standard Operating Procedures. 2004. Seoul: MAF;83.
21. Mitnaul LJ, Matrosovich MN, Castrucci MR, Tuzikov AB, Bovin NV, Kobasa D, Kawaoka Y. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J Virol. 2000. 74:6015–6020.

22. Naeem K, Ullah A, Manvell RJ, Alexander DJ. Avian influenza A subtype H9N2 in poultry in Pakistan. Vet Rec. 1999. 145:560.
23. Ohuchi M, Ohuchi R, Feldmann A, Klenk HD. Regulation of receptor binding affinity of influenza virus hemagglutinin by its carbohydrate moiety. J Virol. 1997. 71:8377–8384.

24. Schulze IT. Effects of glycosylation on the properties and functions of influenza virus hemagglutinin. J Infect Dis. 1997. 176:S24–S28.

26. Swayne DE, Halvorson DA. Saif YM, editor. Influenza. Diseases of Poultry. 2003. 11th ed. Ames: Iowa State Press;135–160.

27. Swayne DE, Senne DA, Beard CW. Swayne DE, Glisson JR, Jackwood MW, Pearson JE, Reed WM, editors. Avian influenza. A Laboratory Manual for the Isolation and Identification of Avian Pathogens. 1998. 4th ed. Pennsylvania: American Association of Avian Pathologists;150–155.
28. Tsuchiya E, Sugawara K, Hongo S, Matsuzaki Y, Muraki Y, Li ZN, Nakamura K. Antigenic structure of the haemagglutinin of human influenza A/H2N2 virus. J Gen Virol. 2001. 82:2475–2484.

29. Vasfi Marandi M, Bozorgmehri Fard MH, Hashemzadeh M. Efficacy of inactivated H9N2 avian influenza vaccine against non-highly pathogenic A/Chicken/Iran/ZMT-173/1999. Arch Razi Institute. 2002. 53:23–32.
30. Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992. 56:152–179.

31. World Organization for Animal Health (OIE). Chapter 2.7.12. Avian influenza. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. 2004. 5th ed. Paris: World Organization for Animal Health (OIE).




 PDF
PDF ePub
ePub Citation
Citation Print
Print






 XML Download
XML Download