Abstract
Twelve Korean infectious bronchitis viruses (IBVs) were isolated in the field from chickens suspected of being carriers of infectious bronchitis between 2001 and 2003. The S1 glycoprotein genes of these IBV isolates were amplified by reverse transcriptase-polymerase chain reaction (RT-PCR) and analyzed by restriction fragment length polymorphism (RFLP) analysis. These Korean IBV isolates were classified into three groups according to their RFLP patterns obtained using the restriction enzyme HaeIII. Half of the twelve isolates were similar to the KM91 RFLP pattern, which is a common pattern in Korea. Three more isolates were related to the Arkansas strain pattern, but with some unique variations. The other three viruses showed variant RFLP patterns. For a comparison with the published sequences for non-Korean IBV strains, amplified PCR products from the twelve isolates were cloned and sequenced. The Korean IBV field isolates had 71.2-99.7% nucleotide sequence homology with each other and 45.9-80.7% nucleotide sequence homology with non-Korean IBV strains. With respect to the deduced amino acid sequence, the Korean IBV isolates had 71.5-99.3% similarity with each other and 44.9-80.3% similarity with non-Korean IBV strains. Phylogenetic tree analysis revealed that some of the IBV isolates appear to belong to a new group, different from the non-Korean IBV strains or from previously isolated Korean IBV strains. Specifically, the new Korean IBV isolates K10217-03, K3-3 and K1255-03 represented a separate group. These findings suggest that the Korean IBVs appear to be continuously evolving.
The avian infectious bronchitis virus (IBV) causes economically important upper respiratory and urogenital tract diseases in chickens, resulting in tracheal rales, sneezing, coughing, reduced weight gain, and reduction of egg production [5]. IBV, the causative agent of infectious bronchitis (IB), belongs to the family Coronaviridae and is found worldwide [5,15]. The genome of IBV consists of a single-stranded sense RNA genome encoding four structural proteins, which are envelope glycoprotein, integral membrane glycoprotein, phosphorylated nucleocapsid protein, and spike (S) glycoprotein [15,22].
The S glycoprotein is cleaved post-translationally by cellular proteases into the S1 and S2 subunits [4]. The globular S1 subunit forms the tip of a spike, extending outward, whereas the S2 subunit anchors the S1 moiety to the viral membrane [1]. The S1 subunit is involved in viral infectivity, virus-neutralizing epitopes, serotype-specific sequences, and hemagglutinin activity [1-3,11,12].
Different serotypes and subtypes of IBV have been reported worldwide, including in Korea [8,9,14,17-19,21]. Various serotypes are thought to develop by nucleotide insertions, deletions, point mutations, and by RNA recombination in the S1 subunit [10,13,23,24]. Since IBV was first reported in Korea in 1986 and nephropathogenic IBV was recognized in 1990, a variety of serotypes of IBV have been reported in Korea [17]. Some of these IBV isolates exhibit variant patterns that distinguish them from each other and from non-Korean IBV isolates in analysis by reverse transcriptase-polymerase chain reaction-restriction fragment length polymorphism (RT-PCR-RFLP) [17].
The objective of this study was genetic characterization of recent IBV isolates in Korea. The S1 glycoprotein genes of the Korean IBVs were amplified by RT-PCR. Amplified S1 genes were classified by RFLP analysis and cloned, sequenced and compared to other non-Korean published IBV sequences. By RT-PCR-RFLP and phylogenetic tree analysis, recent Korean IBV isolates were classified at the genetic level into three distinct groups, two of which included only indigenous Korean IBV isolates and one of which represented a new phylogenetic group.
All procedures for RNA isolation and reverse transcriptase-polymerase chain reaction (RT-PCR) were previously described [14]. Viral RNA was isolated and purified from allantoic fluid collected from embryonated SPF chicken eggs inoculated with IBV isolates using the QIAamp MinElute Virus Spin Kit (QIAGEN, USA).
Amplification of the S1 gene by RT-PCR was performed using one of the forward primers S1OLIGO5' (5'TGA AAACTGAACAAAAGA3') or NEWS1OLIGO5' (5'TG AAAACTGAACAAAAGAC3') together with one of the reverse primers S1OLIGO3' (CTAAACTAACATAAGGG C3') or degenerate3'-2 (5'CCATAAGTAACATAAGGG CAA3') [14,16]. The RT reaction for synthesis of cDNA consisted of purified RNA, 20 pmol S1 OLIGO3' or degenerate 3'-2 primer and RT PreMix AccuPower RT PreMix (Bioneer, Korea). The mixture was incubated at 42℃ for 60 min, and then heated for 5 min at 94℃ to stop the reaction. For the PCR reaction, 20 pmol each primer and cDNA were added to the AccuPower PCR PreMix (Bioneer, Korea). The PCR was performed with 35 cycles of denaturation at 94℃ for 90 sec, annealing at 45℃ for 30 sec, polymerization at 72℃ for 90 sec and a final polymerization step at 72℃ for 10 min. The PCR products were analyzed on a 1.0% agarose gel. The predicted size of the PCR products was about 1,720 bp [14,16].
Amplification products were excised from an agarose gel and purified using the GENECLEAN Turbo Kit (Qbiogene, USA). The restriction enzyme HaeIII was used to digest the S1 gene amplification product as previously described [14,17], and the resulting RFLP patterns were observed after electrophoresis on a 2% agarose gel.
Twelve IBV isolates selected after RFLP analysis were sequenced. PCR products were cut from 1% agarose gels and purified using the GENECLEAN Turbo Kit (Qbiogene, USA). Purified PCR products were then cloned into the pGEM-T Easy Vector (Promega, USA) and transformed into JM 109 competent cells (Promega, USA). The cells carrying recombinant plasmids were selected on Luria-Bertani agar plates containing ampicillin, X-gal and IPTG, and plasmid DNA for sequencing was prepared using the E.Z.N.A plasmid miniprep Kit I (Omega Bio-tec, USA). Sequencing was performed with T7 and SP6 promoter primers using an ABI PRISM 3700 DNA Analyzer (Applied Biosystems, USA). For each IBV isolate, two or three independent clones originating from different PCR reactions were sequenced in order to guard against the possibility of errors arising during the RT-PCR or cloning steps.
Nucleotide sequence data were compiled and analyzed using the Clustal V method in MegAlign software (DNA Star, USA). A phylogenetic tree for the S1 glycoprotein was generated using the maximum parsimony method with 100 bootstrap replicates in a heuristic search with the PAUP 4.0 software program (Sinauer Associates, USA).
The sequence data for the S1 gene reported in this paper were added to the GenBank database (Table 1). Sequences used for comparison or phylogenetic analysis in this study were obtained from the following GenBank database accession numbers: Arkansas 99 (M85244), Beaudette (X 02342), Connecticut (L18990), DE072 (U77298), Gray (L14069), H120 (M21970), Mass 41 (X04722), KM91 (EF621369), K069-01 (AY257061), K281-01 (AY257062), and K774-01 (AY257065).
The initial classification of the IBV isolates was accomplished by RT-PCR-RFLP analysis. Twelve Korean IBVs were separated into four groups by RFLP analysis (Fig. 1). Six of the twelve isolates (K748-01, K044-02, K058-02, K234-02, K117-02, and K514-03) had the same RFLP pattern as the IBV KM91 strain, which was isolated in 1991 in Korea [17]. The RFLP pattern for isolate K434-01 corresponded to an IBV Arkansas strain. The RFLP patterns of the K2-6 and K545-02 isolates were related to the Arkansas strain but had unique patterns [14,17]. Three of the Korean IBVs (K10217-03, K3-3 and K1255-03) generated an identical but variant RFLP pattern.
The S1 gene of the 12 Korean IBV strains was sequenced to characterize the isolates. The nucleotide and deduced amino acid sequences of those IBV isolates were determined and compared with the sequences of published non-Korean IBV strains (Table 2, Fig. 2).
Korean IBVs had nucleotide sequence identities of between 71.2% (K545-02 and K3-3) and 99.7% (K748-01 and K117-02) with each other and between 45.9% (DE072 and K2-6) and 80.7% (H120 and K044-02) with non-Korean IBVs. Korean IBVs had amino acid sequence similarities of between 71.5% (K545-02 and K3-3) and 99.3% (K748-01 and K117-02) with each other and between 44.9% (DE072 and K2-6) and 80.3% (BEAU and K044-02) with non-Korean IBVs.
The deduced amino acid sequences of Korean IBVs were aligned with the sequences of published Korean and non-Korean strains (Fig. 2). Most variations were observed among residues 53-96, 115-163 and 268-398 (numbering is with reference to the Mass41 strain).
A phylogenetic tree was constructed from the nucleotides and deduced amino acid sequences of the S1 glycoprotein genes of the Korean and non-Korean IBVs (Fig. 3). The twelve Korean IBVs were grouped into three distinct clusters. Recent IBV isolates K10207-03, K3-3 and K1255-03 formed the first independent branch. The six additional IBVs K514-03, K044-02, K058-02, K234-02, K117-02, and K748-01 formed the second group, along with the K069-01 and K774-01 strains that were grouped into the KM91 type previously [17]. Finally, the K2-6, K434-01 and K545-02 isolates formed a third group that was related to the IBV Ark99 and Gray strains.
Although a Mass-type live attenuated vaccine and inactivated vaccine have been widely used to control IB, the disease has continued to be a problem in Korea. Twelve Korean IBVs were analyzed in this study, first by RT-PCR-RFLP and then by nucleotide sequencing of the S1 glycoprotein gene.
The Korean IBV field isolates were studied between 1986 and 1997 and were characterized using RT-PCR-RFLP analysis and pathogenicity testing, but the sequences of those viruses were not reported [21]. According to those prior analyses, the KM91 type is the most common or representative genotype III among the five genotypes. KM91 yielded distinct RFLP patterns in the PCR-RFLP analysis using the restriction enzymes Hae III, Eco RI and Bam HI. For the pathogenicity testing, the isolate KM91 was associated with 50% mortality, severe nephritis and renal urate deposits in the kidneys of infected chicks, whereas the other strains merely caused respiratory distress one to two days after inoculation [21]. The H120 vaccine could not protect the chicks against the challenge with the KM91 isolate [21]. In the RT-PCR-RFLP analysis of the recent IBV isolates, 10 of 15 IBVs produced RFLP patterns corresponding to the IBV KM91 strain [17]. Therefore, IBV KM91 seems to be the major IBV in Korea.
In this study, half of the 12 Korean IBV isolates (K748-01, K044-02, K058-02, K117-02, K234-02, and K514-03) sequenced were classified as belonging to the KM91 type by RFLP analysis, and these had 71.2% to 99.7% nucleotide sequence identity and 71.5% to 99.3% amino acid sequence similarity with each other. Although these IBVs exhibited identical RFLP patterns, differences in genetic composition might still exist that could affect their behavior under field conditions.
In the phylogenetic tree, the Korean IBV isolates examined formed three different groups. Half of the 12 Korean IBVs (K748-01, K044-02, K058-02, K117-02, K234-02, and K514-03) were classified into the IBV KM91 type, consistent with the result obtained by RT-PCR-RFLP analysis [21].
The three IBVs K10217-03, K3-3 and K1255-03 recently isolated in Korea formed a distinct cluster, which was related to the KM91 type. They shared between 83.3% to 85.2% amino acid sequence similarity with the KM91 type IBVs, a higher similarity with KM91 than non-Korean and some other Korean IBVs. These three IBVs shared a unique RFLP pattern, however, which had not been previously reported. Therefore, the Korean IBV K10217-03, K3-3 and K1255-03 isolates seem to represent a new Korean IBV variant. Further characterization of these IBV isolates by virus-neutralization testing is warranted. The remaining isolates K434-01, K545-02, and K2-6 were clustered into the Ark99 group, although K545-02 and K2-6 isolates were not classified into the Arkansas type by PCR-RFLP analysis. They showed various RFLP patterns. Either these strains generate two or more RFLP patterns in spite of belonging to the same group in phylogenetic tree analysis, or they may in fact consist of a mixture of two different kinds of viruses. Distinguishing between these possibilities will require further characterization using cloned viruses.
The evolution of IBV appears to be influenced by many factors, such as the use of multiple strains for vaccination, the population density and the host immune status [6]. In addition, transcription of IBV's RNA genome has a high error rate [15,20]. Widespread uses of various vaccines made from heterologous IBVs in the field may also exert pressure resulting in the increase of new genetic variants in Korea.
In summary, 12 Korean field IBVs were isolated and found to differ from published non-Korean IBV strains in both nucleotide and amino acid sequences. Phylogenetic tree analysis revealed three distinct groups, one of which has not been previously reported. It appears that the IBVs are continuously evolving in Korea. Therefore, the indigenous IBV isolates should be considered as the initial candidates for protection against current IBV infections in chickens. Virus-neutralization and cross-challenge tests will be needed for further characterization of the new IBVs before selecting strains for a vaccine.
Figures and Tables
Fig. 1
RFLP patterns of the PCR-amplified S1 glycoprotein genes from representative Korean IBV isolates digested with restriction enzyme Hae III. Lanes 1: 100 bp plus DNA ladder; Lane 2: K044-02; Lane 3: K434-01; Lane 4: K 3-3; Lane 5: K2-6; Lane 6: K545-02.
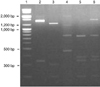
Fig. 2
The deduced amino acid sequences of the S1 glycoprotein gene of 13 Korean IBV isolates and six published non-Korean IBV strains. The dashes (-) indicate regions where the sequences are identical to those of K748-01. Deletions within the sequences are shown with asterisks (*).

Fig. 3
Phylogenetic relationship based on the deduced amino acid sequences of the S1 glycoprotein of the 12 Korean IBV field isolates (K434-01, K748-01, K058-02, K044-02, K117-02, K234-02, K545-02, K514-03, K10217-03, K1255-03, K3-3, K3-3) and non-Korean IBV strains generated by maximum parsimony method with heuristic search and 100 bootstrap replicates. The length of each branch represents the number of amino acid changes between sequences.

Table 1
History of Korean IBV field isolates, RFLP patterns and GenBank accession numbers

*K = kidney, T = trachea, CT = cecal tonsil. †B = broiler, BB = broiler breeder. ‡KM91 was the representative isolate of Korean IBV isolates determined as genotype III which showed a distinct RFLP pattern in PCR-RFLP analysis [21]. §Unidentified.
Acknowledgments
This work was supported by the Agricultural R&D Promotion Center, Korea (Grant No. 0903006-1), and Institute of Veterinary Science, Kangwon National University, Korea.
References
1. Cavanagh D. Coronavirus IBV: structural characterization of the spike protein. J Gen Virol. 1983. 64:2577–2583.

2. Cavanagh D, Davis PJ. Coronavirus IBV: removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J Gen Virol. 1986. 67:1443–1448.

3. Cavanagh D, Davis PJ, Darbyshire JH, Peters RW. Coronavirus IBV: virus retaining spike glycopolypeptide S2 but not S1 is unable to induce virus-neutralizing or hemagglutination-inhibiting antibody, or induce chicken tracheal protection. J Gen Virol. 1986. 67:1435–1442.

4. Cavanagh D, Davis PJ, Pappin D, Binns MM, Boursnell M, Brown T. Coronavirus IBV: partial amino terminal sequencing of spike polypeptide S2 identifies the sequence Arg-Arg-Phe-Arg-Arg at the cleavage site of the spike precursor propolypeptide of IBV strains Beaudette and M41. Virus Res. 1986. 4:133–143.

5. Cavanagh D, Naqi SA. Calnek BW, Barnes HJ, Beard CW, McCougald LR, Saif YM, editors. Infectious bursal disease. Diseases of Poultry. 1997. 10th ed. Ames: Iowa state University Press;511–526.
6. Domingo E, Martínez-Salas E, Sobrino F, de la Torre JC, Portela A, Ortín J, López-Galindez C, Pérez-Breña P, Villanueva N, Nájera R. The quasispecies (extremely heterogeneous) nature of viral RNA genome population; biological relevance-a review. Gene. 1985. 40:1–8.

7. Gelb J Jr, Jackwood MW. Swayne DE, editor. Infectious bronchitis. A Laboratory Manual for the Isolation and Identification of Avian Pathogens. 1998. 4th ed. Georgia: American Association of Avian Pathologists;169–173.
8. Gharaibeh SM. Infectious bronchitis virus serotypes in poultry flocks in Jordan. Prev Vet Med. 2007. 78:317–324.

9. Jackwood MW, Hilt DA, Lee CW, Kwon HM, Callison SA, Moore KM, Moscoso H, Sellers H, Thayer S. Data from 11 years of molecular typing infectious bronchitis virus field isolates. Avian Dis. 2005. 49:614–618.

10. Jia W, Karaca K, Parrish CR, Naqi SA. A novel variant of infectious bronchitis virus resulting from recombination among three different strains. Arch Virol. 1995. 140:259–271.

11. Karaca K, Naqi S, Gelb J Jr. Production and characterization of monoclonal antibodies to three infectious bronchitis virus serotypes. Avian Dis. 1992. 36:903–915.

12. Koch G, Hartog L, Kant A, van Roozelaar DJ. Antigenic domains on the peplomer protein of avian infectious bronchitis virus: correlation with biological functions. J Gen Virol. 1990. 71:1929–1935.

13. Kusters JG, Niesters HGM, Bleumink-Pluym NMC, Davelaar FG, Horzinek MC, van der Zeijist BAM. Molecular epidemiology of infectious bronchitis virus in The Netherlands. J Gen Virol. 1987. 68:343–352.

14. Kwon HM, Jackwood MW, Gelb J Jr. Differentiation of infectious bronchitis virus serotypes using polymerase chain reaction and restriction fragment length polymorphism analysis. Avian Dis. 1993. 37:194–202.

16. Lee CW, Hilt DA, Jackwood MW. Redesign of primer and application of the reverse transcriptase-polymerase chain reaction and restriction fragment length polymorphism test to the DE072 strain of infectious bronchitis virus. Avian Dis. 2000. 44:650–654.

17. Lee SK, Sung HW, Kwon HM. S1 glycoprotein gene analysis of infectious bronchitis viruses isolated in Korea. Arch Virol. 2004. 149:481–494.

18. Liu SW, Zhang QX, Chen JD, Han ZX, Liu X, Feng L, Shao YH, Rong JG, Kong XG, Tong GZ. Genetic diversity of avian infectious bronchitis coronavirus strains isolated in China between 1995 and 2004. Arch Virol. 2006. 151:1133–1148.

19. Mondal SP, Cardona CJ. Genotypic and phenotypic characterization of the California 99 (Cal99) variant of infectious bronchitis virus. Virus Genes. 2007. 34:327–341.

20. Sawicki SG, Sawicki DL. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990. 64:1050–1056.

21. Song CS, Lee YJ, Kim JH, Sung HW, Lee CW, Izumiya Y, Miyazawa T, Jang HK, Mikami T. Epidemiology classification of infectious bronchitis virus isolated in Korean between 1986 and 1997. Avian Pathol. 1998. 27:409–416.

22. Stern DF, Sefton BM. Coronavirus proteins: biogenesis of avian infectious bronchitis virus virion proteins. J Virol. 1982. 44:794–803.

23. Wang L, Junker D, Collisson EW. Evidence of natural recombination within the S1 gene of infectious bronchitis virus. Virology. 1993. 192:710–716.

24. Wang L, Junker D, Hock L, Ebiary E, Collisson EW. Evolutionary implications of genetic variations in the S1 gene of the infectious bronchitis virus. Virus Res. 1994. 34:327–338.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download