The susceptibility to prion diseases in humans and sheep is strongly associated with different genotypes of the prion protein gene (PRNP) [1,2]. Chronic wasting disease (CWD), which is a form of transmissible spongiform encephalopathy, principally affects mule deer, white-tailed deer, and Rocky Mountain elk [9]. The specific PRNP allele patterns found in these species significantly influence their susceptibility to CWD as well as the progression of the disease. Although the PRNP of the sika deer encodes almost identical amino acids to those of CWD-susceptible deer species there is still no definitive report on CWD-infected sika deer. Therefore, we are currently unable to determine the correlation between a specific allele of the PRNP and the CWD susceptibility of sika deer. Instead, single-nucleotide polymorphisms (SNPs) of PRNP have been identified in both Chinese and Japanese domestic sika deer [5,6]. This study characterized three SNPs in the PRNP of sika deer (Cervus nippon laiouanus) reared in Korea, one of which had not been reported previously.
Blood samples from 33 sika deer were provided by the Seoul Grand Park Zoo (Korea). The genomic DNA was isolated from the blood using a DNeasy Blood and Tissue kit (Qiagene, USA), in accordance with the manufacturer's instructions. The DNA samples were used to prepare the templates for the PCR amplification of sika deer PRNP. The primers used in the amplification of 771 bp of the sika deer PRNP were designed based on the Rocky Mountain elk PRNP (AF016227). The primers harbored the following DNA sequences: forward 5'-ATG GTG AAA AGC CAC ATA GGC-3' and reverse 5'-CTA TCC TAC TAT GAG AAA AAT G-3'. PCR was carried out in a total reaction volume of 50 ml. The PCR conditions were as follows: initial denaturation for 5min at 94℃, 30 cycles of denaturation at 94℃ for 45 sec, annealing at 58℃ for 45 sec and extension at 72℃ for 90 sec. A final 5-min extension step was completed at 72℃. The PCR products were analyzed by electrophoresis using 1.5% agarose gel before purification with a PCR purification kit (Qiagen, USA). The purified DNA samples were submitted for DNA sequencing. The DNA sequencing reactions were carried out in an MJ Research PTC-225 Peltier Thermal Cycler, using an ABI PRISM BigDye Terminator Cycle Sequencing Kit (ABI, USA). Each of the PCR products was sequenced in both directions using the same primers used for PCR.
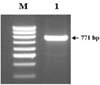
The sika deer PRNP, which is composed of 771 bp, was amplified successfully from the blood-derived genomic DNA by PCR (Fig. 1). The DNA sequencing result confirmed the identity of the PCR product as the PRNP of sika deer. The electropherograms of the DNA sequencing results of each sample were analyzed to determine the SNPs. A total of three SNPs were identified in the sika deer PRNP at codons 100, 136 and 226 (Figs. 2-4). Their sequences were deposited at the GenBank database (DQ358970 and EF057409). The SNP (A→G at nucleotide 298) detected at codon 100 was found to be a novel SNP in the sika deer PRNP, which resulted in a serine to glycine amino acid change (Fig. 2). The allele frequencies of the polymorphism were 96.97 and 3.03% for A and G, respectively (Table 1). Two SNPs located at codons 136 and 226 have previously been reported in the Chinese domestic sika deer [6]. The SNP (T→C at nucleotide 408) at codon 136 was found to be a silent mutation, which did not induce any amino acid substitutions (Fig. 3), as previously reported in studies of the sika deer inhabiting China and Japan [5,6]. The allele frequencies of that polymorphism were 59.09 and 40.91% for T and C, respectively (Table 1). However, the SNP (C→G at nucleotide 676) located at codon 226 induced amino acid changes from glycine to glutamic acid (Fig. 4). The allele frequencies of that polymorphism were 48.48 and 51.52% for C and G, respectively (Table 1).
The SNPs in the sika deer PRNP have been identified in Chinese and Japanese domestic sika deer at nucleotides 408 and 676, and 63, 255 and 408, respectively [5,6]. The allele frequencies detected at codons 136 (nucleotide position 408) and 226 (nucleotide position 676) in the PRNP of Korean domestic sika deer were only slightly different from those reported in Chinese domestic sika deer [6]. While the Chinese domestic sika deer had 55% for the C allele of the 408 SNP and 55% for the G allele of the 676 SNP, the Korean domestic sika deer showed 40.91% and 51.52%, respectively. However, the polymorphisms identified at nucleotides 63 and 225 in the PRNP of the Japanese domestic sika deer could not be identified in this study [5]. These discrepancies might be the result of a different subspecies of sika deer being studied in each country or the relatively small sample numbers analyzed in this study. In addition, the amino acid residues, 95Q, 96G, 116A, 132M, 225S, and 226Q, which were most commonly detected in the PRNP alleles of CWD-susceptible mule deer, white-tailed deer, and elk [3,4,7,8] were also identified in the sika deer (data not shown). More CWD-affected Rocky Mountain elk had a 132M homozygous genotype than the uninfected animals [7]. The alleles, 95Q, 96G, 116A and 226Q, were reported to be over-represented in the CWD-susceptible white-tailed deer [4,8]. In addition, mule deer with a 225S homozygous genotype had a higher susceptibility to CWD than the animals with a heterozygous genotype at codon 225 [3]. However, we could not determine the precise association of the polymorphisms and amino acids addressed in this study with CWD-susceptibility of sika deer. Therefore, more study will be needed to address the issue of CWD-susceptibility in sika deer.




 PDF
PDF ePub
ePub Citation
Citation Print
Print






 XML Download
XML Download