Abstract
The present study was carried out to genotypically characterize Staphylococcus aureus (S. aureus) isolated from bovine mastitis cases. A total of 37 strains of S. aureus were isolated during processing of 552 milk samples from 140 cows. The S. aureus strains were characterized phenotypically, and were further characterized genotypically by polymerase chain reaction using oligonucleotide primers that amplified genes encoding coagulase (coa), clumping factor (clfA), thermonuclease (nuc), enterotoxin A (entA), and the gene segments encoding the immunoglobulin G binding region and the X region of protein A gene spa. All of the isolates yielded an amplicon with a size of approximately 1,042 bp of the clfA gene. The amplification of the polymorphic spa gene segment encoding the immunoglobulin G binding region was observed in 34 isolates and X-region binding was detected in 26 isolates. Amplification of the coa gene yielded three different products in 20, 10, and 7 isolates. The amplification of the thermonuclease gene, nuc, was observed in 36 out of 37 isolates. All of the samples were negative for the entA gene. The phenotypic and genotypic findings of the present strategies might provide an understanding of the distribution of the prevalent S. aureus clones among bovine mastitis isolates, and might aid in the development of steps to control S. aureus infections in dairy herds.
Staphylococcus aureus (S. aureus) is one of the common causes of subclinical mastitis worldwide, which is of economic importance to the dairy industry [13]. Raw milk is a potential source of S. aureus in milk and milk products, especially in the case of defective pasteurization. The main reservoir of S. aureus seems to be the infected quarter. Molecular epidemiological analysis of the bovine S. aureus population suggested that a small number of clonal types were responsible for most infections, and that strains had a broad geographic distribution [7,11,15].
S. aureus has a capacity to produce a large number of putative virulence factors [6,8,9]. Some of these factors may be of more importance than others in different diseases or at different stages of the pathogenesis of particular infections, as not all factors are produced by each strain. At present, nothing has been reported about the occurrence of these virulence factors among S. aureus isolates from India, and about the possible distribution of single S. aureus clones as causative agents of bovine mastitis at various farms. The present study was conducted to genotypically characterize S. aureus isolates in milk samples from cows with subclinical mastitis.
A total of 552 quarter milk samples were collected from 140 cows selected randomly from 8 farms in the Vidarbha region of Central India. Samples were collected over a two month period. The samples were tested by the California mastitis test (CMT) for subclinical mastitis, and were graded as negative, trace, weak, distinct, or strongly positive [14]. Isolation of Staphylococcus was attempted from the CMT positive milk samples.
Chromosomal DNA of the isolates was prepared as described by Wilson [19] with some modification. In brief, bacteria were grown in brain heart infusion broth for 24 h at 37℃. The cultures were centrifuged at 4℃ at 8,000 g for 10 min. The pellet was suspended in TE buffer (200 µl) (10 mM Tris HCl + 1 mM EDTA, pH 8.0) with lysozyme (10 mg/ml) and incubated at 37℃ for 2 h. The bacteria was lysed with 10% SDS and proteinase K (10 mg/ml), and was incubated at 65℃ for 30 min. The denatured protein, cell wall debris, and polysaccharides were eliminated by the addition of 5M NaCl and CTAB/NaCl (10% hexadecyl trimethyl ammonium bromide in 0.7M NaCl) and incubated for 30 min at 65℃. DNA was purified by extraction with phenol: chloroform (1 : 1) and chloroform: isoamyl alcohol (24 : 1). DNA was precipitated with isopropanol and sodium acetate (3 M) solutions, washed in 70% ethanol, and suspended in 50 µl of TE buffer.
The virulence determinants investigated using the oligonucleotide primers included the genes encoding coagulase (coa), clumping factor (clfA), the IgG-binding region and the X-region of protein A (spa), enterotoxin A (entA), and thermonuclease (nuc). For all the genes, reaction mixtures (25 µl) included 2 µl template DNA, 10 × PCR buffer (Sigma Aldrich, USA), 25 mM MgCl2, 200 µM of the four dNTPs, 10 pmol of each of the 2 primers (Bangalore Genei, India), and 1U Taq DNA polymerase (Sigma Aldrich, USA).
In the present study, the amplification parameters and primer sequences described by Straub et al. [18] were used (Table 1). The amplification of genes was carried out with thermocycler (Thermo Hybaid, USA).
Amplified products were separated by agarose gel electrophoresis (1.5% agarose containing 0.5 mg ethidium bromide in 0.5 × Tris-EDTA electrophoresis buffer) at 5 V/cm for 2 h and photographed under UV illumination.
Isolation of Staphylococcus was attempted from CMT positive milk samples. Out of 552 milk samples collected from 140 cows on 8 farms, 501 (90.76%) samples from 134 cows were found to be CMT positive. Of these 268 milk samples, 114 cows harbored Staphylococcus sp. On the basis of cultural and biochemical properties, 37 isolates were identified as S. aureus. All 37 isolates were positive for the tube coagulase test. Others strains were identified as S. intermedius, S. hyicus, coagulase negative staphylococci, and Micrococcus (data not shown).
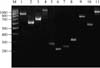
Amplification of the coa gene yielded three different products of 627, 710, and 910 bp for 20, 10, and 7 isolates from 7, 4, and 5 farms, respectively, and gene polymorphism was observed in isolates originating from 5 farms. All of the isolates yielded an amplicon with a size of approximately 1,042 bp of the clfA gene. The amplification of the gene segment encoding the IgG binding region of protein A (spa) revealed a size of 590, 810, and 970 bp in 12, 15, and 7 isolates from 5, 5, and 4 farms, respectively, and gene polymorphism was noted in isolates from 4 farms. The X-region binding of the spa gene produced an amplicon of 220, 253, and 315 bp in 10, 9, and 7 isolates, respectively. The amplification of the extracellular thermonuclease nuc gene produced an amplicon of 279 bp in 36 out of 37 isolates. All of the samples were found to be negative for the entA gene. Amplicons specific to the coa, clfA, nuc, spa IgG binding, and X-region genes are shown in Fig. 1. The genotypic properties of the 37 S. aureus isolates are summarized in Table 2.
S. aureus has been recognized as a pathogen in human and animal infections. In the present study, 37 S. aureus strains isolated from subclinical bovine mastitis cases were identified and further characterized by PCR amplification of various virulence genes encoding clumping factor and coagulase activity, and gene segments encoding the immunoglobulin G-binding region and X-region of protein A and stable thermonuclease activity. Comparable PCR-based detection studies of the virulence genes have been described by other investigators [1,15].
The coa and spa (IgG-binding region and X-region) genes investigated in the present work exhibited typical gene polymorphism. This attribute could be used for the genotypic characterization of single isolates of this species. The spa gene segments encoding the X-repetitive region are known to consist of a variable number of small repeats [5]. It is thought that the spa domain encoding the X-region may serve to extend the N-terminal IgG-binding portion of the protein through the cell wall. It was interesting to note that isolates from the same farm exhibited polymorphism among the coa and spa genes.
The ability of S. aureus to adhere to extracellular matrix proteins is thought to be essential for the colonization and the establishment of infections [5]. S. aureus possesses various adhesion genes, including clfA, fnbA, and cna [16]. PCR analysis of the other virulence genes revealed the nuc and clfA genes in 36 and 37 strains, respectively, of the 37 strains investigated, suggesting an important role of these elements in the pathogenecity of bovine mastitis. However, entA was not present among the strains. In contrast, combined occurrence of enterotoxin genes has been described by other investigators [1,10,17,20].
In the present study, S. aureus isolates from cattle with bovine mastitis were found to differ in their gene patterns. Phenotypic and genotypic characterization might provide a better understanding of the distribution of the prevalent S. aureus clones among bovine mastitis isolates. This can aid in the investigation and control of S. aureus infections in dairy herds.
Figures and Tables
Fig. 1
Amplicons of the genes encoding Staphylococcal coagulase (coa), clumping factor (clfA), thermonuclease (nuc), spa gene X-region, and IgG-binding regions. Lane M: DNA molecular weight marker MBD 13 (Bangalore Genei, India); Lane 1-3: coa; Lane 4: clfA; Lane 5: nuc; Lane 6-8: spa X-region; Lane 9-11: spa IgG-binding region.
Acknowledgments
The authors are thankful to the Dean of Nagpur Veterinary College, Seminary Hills, Nagpur, Maharashtra, India for providing the facilities to conduct this research.
References
1. Akineden Ö, Annemüller C, Hassan AA, Lämmler C, Wolter W, Zschöck M. Toxin genes and other characteristics of Staphylococcus aureus isolates from milk of cows with mastitis. Clin Diagn Lab Immunol. 2001. 8:959–964.

2. Annemüller C, Lämmler C, Zschöck M. Genotyping of Staphylococcus aureus isolated from bovine mastitis. Vet Microbiol. 1999. 69:217–224.
3. Cowan ST, Steel KJ. Manual for the Identification of Medical Bacteria. 1970. London: Cambridge University Press;52–57.
4. Cruickshank R, Duguid JP, Harmoin BP, Swain RHA, editors. The practice of medical microbiology. Medical Microbilogy. 1975. Vol. 2:12th ed. Edinburgh: Churchill Livingstone;356–366.
5. El-Sayed A, Alber J, Lämmler C, Bonner B, Huhn A, Kaleta EF, Zschöck M. PCR-based detection of genes encoding virulence determinants in Staphylococcus aureus from birds. J Vet Med B Infect Dis Vet Public Health. 2005. 52:38–44.

6. Fitzgerald JR, Hartigan PJ, Meaney WJ, Smyth CJ. Molecular population and virulence factor analysis of Staphylococcus aureus from bovine intramammary infection. J Appl Microbiol. 2000. 88:1028–1037.

7. Fitzgerald JR, Meaney WJ, Hartigan PJ, Smyth CJ, Kapur V. Fine-structure molecular epidemiological analysis of Staphylococcus aureus recovered from cows. Epidemiol Infect. 1997. 119:261–269.

8. Foster G, Ross HM, Hutson RA, Collins MD. Staphylococcus lutrae sp. nov., a new coagulase-positive species isolated from otters. Int J Syst Bacteriol. 1997. 47:724–726.

9. Foster SJ. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J Bacteriol. 1995. 177:5723–5725.

10. Jarraud S, Cozon G, Vandenesch F, Bes M, Etienne J, Lina G. Involvement of enterotoxins G and I in staphylococcal toxic shock syndrome and staphylococcal scarlet fever. J Clin Microbiol. 1999. 37:2446–2449.

11. Kapur V, Sischo WM, Greer RS, Whittam TS, Musser JM. Molecular population genetic analysis of Staphylococcus aureus recovered from cows. J Clin Microbiol. 1995. 33:376–380.

12. Lachica RVF, Genigeorgis C, Hoeprich PD. Metachromatic agar-diffusion methods for detecting Staphylococcal nuclease activity. Appl Microbiol. 1971. 21:585–587.

13. Miles H, Lesser W, Sears P. The economic implications of bioengineered mastitis control. J Dairy Sci. 1992. 75:596–605.

14. Radostits OM, Gay CC, Blood DC, Hinchcliff KW. Veterinary Medicine. A Textbook of the Diseases of Cattle, Sheep, Pigs, Goats and Horses. 2003. 9th ed. Philadelphia: Saunders;615.
15. Salasia SIO, Khusnan Z, Lämmler C, Zschöck M. Comparative studies on pheno- and genotypic properties of Staphylococcus aureus isolated from bovine subclinical mastitis in central Java in Indonesia and Hesse in Germany. J Vet Sci. 2004. 5:103–109.

16. Smeltzer MS, Gillaspy AF, Pratt FL Jr, Thames MD, Iandolo JJ. Prevalence and chromosomal map location of Staphylococcus aureus adhesin genes. Gene. 1997. 196:249–259.

17. Stephan R, Annemüller C, Hassan AA, Lämmler C. Characterization of enterotoxigenic Staphylococcus aureus strains isolated from bovine mastitis in north-east Switzerland. Vet Microbiol. 2001. 78:373–382.

18. Straub JA, Hertel C, Hammes WP. A 23S rRNA-targeted polymerase chain reaction-based system for detection of Staphylococcus aureus in meat starter cultures and dairy products. J Food Prot. 1999. 62:1150–1156.

19. Wilson K. Ausuhel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Preparation of genomic DNA from bacteria. Current Protocols in Molecular Biology. 1987. Vol. 1. New York: John Wiley & Sons;2.4.1–2.4.2.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download