Abstract
A total of 22 Salmonella enterica serotype Enteritidis (S. Enteritidis) strains isolated from human and chicken were subjected to DNA fingerprinting by repetitive sequence PCR using ERIC and BOX primers, antibiotic resistance and plasmid patterns. Both ERIC and BOX PCR amplification data revealed a highly genetic homogeneity between isolates from human and chicken except one isolate, which originated from chicken and showed a different DNA band pattern from others. Eleven of 22 S. Enteritidis isolates (50%) were resistant to more than one antibiotics and characterized by 5 resistance patterns. The most common pattern was penicillin resistant (63.6%). Only one isolate from chicken showed a multiple drug resistance patterns to 4 antibiotics. All 22 S. Enteritidis isolates harbored more than two plasmids with eight different plasmid profiles including two to six plasmids with approximate molecular size ranging from 1.9 to 21 kb. A band of 15 kb size was detected in all isolates tested, however, the band sizes smaller than 15 kb were found only in isolates from chicken.
Salmonella enterica serotype Enteritidis (S. Enteritidis) is one of the most important serotypes causing human gastroenteritis outbreaks worldwide during the last few decades. Animals and their products, particularly meat and eggs from chicken, were considered major sources of infections with this pathogen for human [17]. Because of the importance of Salmonella in food-borne diseases, many typing methods have been used to trace the outbreaks to the contaminated source and to elucidate the epidemiology of its infection [7]. Traditional subspecific typing methods include phage typing [14, 18], plasmid profiling [21], multilocus enzyme electrophoresis [5], ribotyping [11] and pulsed field gel electrophoresis (PFGE) [20].
PCR-based fingerprinting is a simple and easily applicable typing method that is potentially available to any laboratory. Families of short repetitive DNA sequences are dispersed throughout the genome of diverse bacterial species [13]. Three families have been studied in more detail including Escherichia coli and Salmonella, namely the 35 to 40 bp repetitive extragenic palindromic (REP) sequence [4], the 124 to 127 bp enterobacterial repetitive intergenic consensus (ERIC) sequence [3] and the 154 bp BOX elements [9]. The actual function of these elements have not been fully known although their involvement in stabilizing mRNA, chromosome organization and binding of DNA polymerase I has been suggested [6]. Widespread distribution of these repetitive DNA elements in the genomes of various microorganisms should enable rapid identification of bacterial species and strains, and were also useful for the analysis of prokaryotic genomes [23]. In this study, 22 S. Enteritidis isolated from human and chicken were fingerprinted by repetitive sequence PCR (rep-PCR) using ERIC and BOX primers to assess genetic relationships between strains of S. Enteritidis from different sources. Also, their antibiotic resistance and plasmid profiles were included.
A total of 22 S. Enteritidis strains were analyzed in this study (Table 1). Ten strains from chickens were isolated from feces of chickens in 3 slaughterhouses, and twelve strains were isolated from fecal samples of 12 food-poisoning outbreaks in Gyeongsang province between 2001 and 2002. All strains were confirmed as S. Enteritidis by conventional biochemical test [12] and serotyped with respect to cell wall (O) and flagella (H) antigens (Difco, USA).
PCR was performed essentially as described by Versalovic et al. [23] with minor modifications. For DNA isolation, 2-3 individual colonies were suspended in 500 ml of distilled water. They were boiled for 5 min, and centrifuged at 8,000 × g. The supernatant was used as DNA and stored at -20℃ until use. Primers included ERIC1R (5'-ATG TAA GCT CCT GGG GAT TCA C-3'), ERIC2 (5'-AAG TAA GTG ACT GGG GTG AGC G-3') and BOXA1R (5'-CTA CGG CAA GGC GAC GCT GAC G-3'). PCR mixtures were prepared in a 25 ml volume containing 2 ml DNA of each isolate, 20 pmol of each primer, 1.25 mM deoxynucleoside triphosphates and 2 U of Taq DNA polymerase (Bioneer, Korea). Amplifications were performed with a UNO II DNA thermal cycler (Biometra, Germany). For the ERIC primers, PCR cycles used were as follows: 1 cycle at 95℃ for 7 min, 30 cycles at 94℃ for 1 min, 52℃ for 1 min and at 65℃ for 8 min. For the ERIC primers, 1 cycle at 95℃ for 7 min was followed by 30 cycles at 94℃ for 1 min, 53℃ for 1 min and at 65℃ for 8 min. After reactions, 10 ml of PCR products were separated on 1.2% agarose gel. The gels were electrophoresed at 4℃ for 10 h at 70 V and stained with ethidium bromide.
Isolates were screened for antimicrobial susceptibility test by an agar diffusion disk method performed on Muller-Hinton agar plates (Difco, USA) [1]. The antibiotics tested were as follows: amikacin (AK; 30 µg), ampicillin (AM; 10 µg), cephalothin (CF; 30 µg), colistin (CL; 10 µg), erythromycin (ER; 15 µg), gentamicin (GM; 10 µg), kanamycin (KM; 30 µg), nalidixic acid (NA; 30 µg), neomycin (NE; 30 µg), penicillin, (PE; 10 U), polymyxin B (PB; 300U), streptomycin (ST; 10 µg), sulfamethoxazole (SX; 300 µg) and tetracycline (TE; 10 µg).
An overnight culture of S. Enteritidis strains in Luria Bertani (Difco, USA) broth at 37℃ was harvested and the cell pellets were subjected to cell lysis, DNA extraction and agaroge gel electrophoresis using plasmid DNA isolation kit (Bioneer, Korea). Band patterns for rep-PCR products and plasmid DNA of each isolate were analyzed using Analysis software (Biometra, Germany), and a tolerance of 5% in the band position was applied. Isolates were considered to have the same electrophoretic profile when their band patterns were identical. Minor differences in band intensity were not considered.
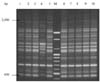
A total of 22 S. Enteritidis strains were analyzed by rep-PCR. DNA fingerprint patterns for S. Enteritidis isolates generated by rep-PCR with ERIC primers showed the identical patterns between isolates from human and chicken sources except one isolate, SC04 (Fig. 1A, B). Each isolate approximately contained between 9 and 10 bands with band sizes ranging from 230 bp to 1,000 bp. Fig. 2 showed the DNA fingerprint patterns of 10 S. Enteritidis isolates from chickens obtained with BOX primer, all showing the same patterns except one isolate (SC04). Twelve isolates from human outbreaks also represented the same patterns with those from chickens (data not shown). Each isolate approximately contained between 14 and 15 bands with band sizes ranging from 450 bp to 2,500 bp. The reproducibility of fragment patterns was stable and reliable when a duplicate analysis of these isolates was performed under identical conditions of template preparation and electrophoresis.
Eleven of 22 S. Enteritidis isolates (50%) were resistant to more than one antibiotics. Also, 11 isolates were characterized by 4 resistance patterns. The most common pattern was PE resistant (63.6%). Only one isolate from chicken showed a multiple drug resistance (MDR) pattern to 4 antibiotics (ST, TE, PE and AM). Two isolates with 3 antibiotics resistant to ST, TE and PE were originated from chicken. Twenty-five and 80% of strains each from human and chicken were sensitive to all antibiotics tested. Table 2 indicates the number and sizes of plasmids from each S. Enteritidis. All 22 S. Enteritidis isolates harbored more than two plasmids. Eight different plasmid profiles, each including two to six plasmids with approximate molecular size ranging from 1.9 to 21 kb, were found. A band of 15 kb size was detected in all isolates tested, but the band sizes of smaller than 15 kb were found only in isolates from chicken.
There have been reports of using rep-PCR fingerprinting technique as an epidemiological tool for several bacterial pathogens. Dombeck et al. [4] reported that the DNA band patterns obtained with BOX primer almost completely separated the human isolates of E. coli from the non-human isolates. Jersek et al. [8] used rep-PCR for typing Listeria monocytogenes strains isolated from humans, animals and foods. Johnson et al. [9] also defined the potential utility of rep-PCR using ERIC primers as a subspecific typing method for Salmonella subspecies. In this study, 22 S. Enteritidis isolates from human and chicken were characterized by rep-PCR methods using ERIC and BOX primers to assess genetic relationships among strains of different sources. Both the ERIC and BOX PCR amplification data revealed the highly genetic homogeneity between isolates from human and chicken except one isolate, which was originated from chicken and showed a different DNA band pattern. We, unfortunately, have no information on epidemiological data on this isolate. It, however, could be concluded that the food-poisoning endemic was caused by a geographically specific S. Enteritidis clone. Also, it might be suggested that the clone was originated from the contaminated chicken products, such as chicken meat and eggs, considering the fact that major source of food-born gastroenteritis in human infections with this pathogen was from animal, particularly chickens [17]. Weigel et al. [24] reported the greater discriminative ability of rep-PCR for genotyping of Salmonella subspecies when compared with PFGE given the equal high reliability of both genotyping methods. Further work on these S. Enteritidis isolates with PFGE tool is underway to compare and elucidate their genetic relationships with PFGE technique.
Considering the marked importance of Salmonella subspecies as food-born pathogen and the worldwide emergence of multi-drug resistant Salmonella strains [10], this study screened the antibiotic resistance profiles of S. Enteritidis isolated from human and chicken. The resistance profiles showed relatively simple but different patterns between them. Nine of 12 isolates (75%) from human were resistant to PE, of which two isolates were resistant to additional AM. Only two isolates from chickens were resistant to ST, TE, and PE, of which one isolate showed an additional AM resistance. Yang et al. [25] reported that 13 of 14 S. Enteritidis isolates from chicken layer between 1995 and 1999 were resistant to SU and that only one isolate was multi-resistant to ST, TE, AM and TE. All isolates, however, were sensitive to SU in this study. Also, results of antimicrobial sensitivity test for isolates over 27,000 cases of human salmonellosis in 2000 in 10 European countries indicated that S. Enteritidis isolates were the most resistant to NA, ST and AM in order [19]. This was different from the results with all isolates from human being sensitive to AM in this study. Rankin and Coyne [15] raised attention of the emergence of multiple antibiotic resistance in S. Enteritidis due to the presence of class I integrons, which have the ability to disseminate the multiple resistance through broad host-range plasmids. It seems that we also need to pay attention to the emergence of these multiple resistant S. Enteritidis isolates in human and animal outbreaks.
The plasmid profile has been used in epidemiological studies of S. Enteritidis [16]. A total of eight plasmid patterns were obtained in this study. The molecular size of the plasmids ranged from 1.9 to 21kb. The band numbers for these patterns were from 2 to maximum of 6. All S. Enteritidis isolates harbored a 15kb size band. Bichler et al. [2] have reported the presence of a 54-57 kb S. Enteritidis serotype-specific plasmid (SSP), which was not detected in this study. It should be noted that considerable size errors could be generated from different laboratories due to the plasmid methodology [22]. It was, however, notable that band sizes of smaller 15kb were detected in 4 of 10 isolates from chicken, which were not found in those from human. Further work will be needed to find whether these unique plasmid bands can probably be used as epidemiological markers to trace the source of S. Enteritidis infection.
Figures and Tables
Fig. 1
DNA fingerprint patterns of S. Enteritidis strains from human (A) and chicken (B) by rep-PCR with the ERIC primers. (A) M, 100 bp ladder; Lane 1-12, SH01-SH12 in order. (B) M, 100 bp ladder; Lane 1-10, SC01-SC10 in order.

Fig. 2
DNA fingerprint patterns of 10 S. Enteritidis strains from chicken by rep-PCR with the BOX primer. M, 100 bp ladder; Lane 1-10, SC01-SC10 in order.
References
1. Bauer AW, Kirby WM, Sherris JC. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966. 45:493–496.

2. Bichler LA, Nagaraja KV, Pomeroy BS. Plasmid diversity in Salmonella enteritidis of animal, poultry and human origin. J Food Prot. 1994. 57:4–11.

3. Burr MD, Josephson KL, Pepper IL. An evaluation of ERIC PCR and AP PCR for fingerprinting for discriminating Salmonella serotype. Lett Appl Microbiol. 1998. 27:24–30.

4. Dombek PE, Johnson LK, Zimmerley ST, Sadowsky MJ. Use of repetitive DNA sequences and the PCR to differentiate Eschrihia coli isolates from human and animal sources. Appl Environ Microbiol. 2000. 66:2572–2577.

6. Gilson E, Perrin D, Hofnung M. DNA polymerase I and a protein complex bind specifically to E. coli palindromic unit highly repetitive DNA: implications for bacterial chromosome organization. Nucleic Acids Res. 1990. 18:3941–3952.

7. Helmuth R, Schroeter A. Molecular typing methods for S. enteritidis. Int J food Micriobiol. 1994. 21:69–77.
8. Jerkek B, Gilot P, Gubina M, Klun N, Mehle J, Tcherneva E, Rijpens N, Herman L. Typing of Listeria monocytogenes strains by repetitive element sequence-based PCR. J Clin Microbiol. 1999. 37:103–109.

9. Johnson JR, Clabots C, Azar M, Boxrud DJ, Besser JM, Thurn JR. Molecular analysis of a hospital cafeteria-associated salmonellosis outbreak using modified repetitive element PCR fingerprinting. J Clin Microbiol. 2001. 39:3452–3460.

10. Kruse H. Globalization of food supply-food safety implications. Special regional requirements: future concerns. Food Control. 1999. 10:315–320.

11. Landeras E, Gonzalez-Hevia MA, Alzugaray R, Mendoza MC. Epidemiological differentiation of pathogenic strains of Salmonella enteritidis by ribotyping. J Clin Microbiol. 1996. 34:2294–2296.

12. Lennette EH, Balows A, Hausler WJ, Shadomy HJ. Manual of Clinical Microbiology. 1985. 4th ed. Washington D.C: Americal Society for Micribiology;1012–1097.
13. Lupski JR, Weinstock GM. Short, interspersed repetitive DNA sequences in prokaryotic genomes. J Bacteriol. 1992. 174:4525–4529.

14. Olsen JE, Skove MN, Threlfall EJ, Brown DJ. Clonal lines of Salmonella enterica serotype Enteritidis documented by IS200-, ribo-, pulsed-field gel electrophoresis and RFLP typing. J Med Microbiol. 1994. 40:15–22.

15. Rankin SC, Coyne MJ. Multiple antibiotic resistance in Salmonella enterica serotype enteritidis. Lancet. 1998. 351:1740.

16. Ridley AM, Threlfall EJ, Rowe B. Genotypic characterization of Salmonella enteritidis phage types by plasmid analysis, ribotyping, and pulsed-field gel electrophoresis. J Clin Microbiol. 1998. 36:2314–2321.

17. Rodrigue DC, Taux RV, Rowe B. International increase in Salmonella enteridis: a new endemic? Epidemiol Infection. 1990. 105:21–27.
18. Terajima J, Nakamura A, wadanabe H. Epidemiological analysis of Salmonella enterica Enteritidis isolates in Japan by phage typing and pulsed field gel electrophoresis. Epideiol Infect. 1998. 120:223–229.

19. Threlfall EJ, Fisher IST, Berghold C, Gerner-Smidt P, Tschape H, Cormican M, Luzzi I, Schnieder F, Wannet W, Machado J, Edwards G. Antimicrobial drug resistance in isolates of Salmonella enterica from cases of salmonellosis in humans in Europe in 2000: results of international multi-center surveillance. Euro Surveill. 2003. 8:41–45.

20. Thong KL, Puthucheary S, Pang T. Outbreak of Salmonella enteritidis gastroenteritidis: Investigation by pulsed-field gel electrophoresis. Int J Infect Dis. 1998. 2:159–163.

21. Threlfall EJ, Hampton MD, Chart H, Rowe B. Use of plasmid profile for surveillance of Salmonella enteritidis phage type 4 from humans, poultry and eggs. Epidemiol Infect. 1994. 112:25–31.

22. Towner KJ, Cockayne A. Molecular Methods for Microbial Identification and Typing. 1993. London: Chapman & Hall;28–63.
23. Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acid Res. 1991. 19:6823–6831.

24. Weigel RM, Qiao B, Teferedegne B, Suh DK, Barber DA, Isaacson RE, White BA. Comparison of pulsed field gel electrophoresis and repetitive sequence polymerase chain reaction as genotyping methods for detection of genetic diversity and inferring transmission of Salmonella. Vet Microbiol. 2004. 100:205–217.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download