Abstract
Toxoplasma gondii is a persistent protozoan parasite capable of infecting almost any warm-blooded vertebrates. SAG1 (p30) is the prototypic member of a superfamily of surface antigens called SRS (SAG1-related sequence). It constitutes the most abundant and predominant antigen. In this paper the primary structure of mature SAG1 gene of an Indonesian T. gondii isolate is described and sequence comparison is made with published sequence data of 7 other strains or isolates. Sequence comparison indicated that SAG1 is highly conserved through evolution and despite parasite spreading world-wide. Sequences may be divided into two major families, independent of the strain/isolate geographic origin. Variations were mainly localized at the C-terminal half or domain 2 and some clustered in restricted areas. Sequence comparison allowed us to define the Indonesian isolate as genuine virulent RH strain. A phylogenetic tree of Toxoplasma strains/isolates was constructed based on SAG1.
Toxoplasmosis is a widely prevalent zoonosis in humans and warm-blooded animals world-wide, due to the tissue cyst-forming coccidium, Toxoplasma gondii. T. gondii is an obligate, intracellular parasite which belongs to the phylum Apicomplexa, a large group of mostly intracellular parasites that includes some deadly pathogens of humans and livestock. While toxoplasmosis is usually innocuous or asymptomatic in most individuals, infection with T. gondii during pregnancy may lead to severe, if not fatal, infection of the fetus [16]. In immunocompromized patients, T. gondii has emerged as an important opportunistic infectious pathogen [17].
T. gondii is one of the most successful protozoan parasites. Transmission of the parasite occurs by ingestion of oocysts shed from feline feces, by ingestion of cysts from chronically infected tissues, or by vertical transmission [16]. The parasite normally divides asexually to yield a haploid form that can infect virtually any vertebrate. However it also has a well defined sexual cycle that occurs exclusively in cats [13]. Felids, domestic and wild, are the only known definitive hosts.
Following initial attachment to host cells, T. gondii develops in a parasitophorous vacuole, that does not fuse with any cell compartment and in which the parasite resides and replicates [5]. An important repertoire of structurally related, yet antigenically distinct surface proteins, called SAG1-related sequence (SRS) proteins, is the key to the success for parasite entry into host cells. This superfamily comprises at least 20 homologous proteins and SAG1 (p30) is the prototypic member [10]. The ability of Toxoplasma to enter and infect a broad spectrum of cell types and hosts may be explained by the function of the SRS family that provides a redundant system of receptors for interaction.
Invasion of host cells requires actin-based motility of the parasite rather than actin-driven internalization by the host cell machinery [4]. This mechanism facilitates parasite migration across cellular barriers and allows dissemination within tissues. Nonviable internalization, for example when opsonized parasites are taken up by phagocytes, leads to internalization in a phagosome and killing of the parasite. Only active invasion leads to parasite development [5].
A large amount of evidence of the essential role of SAG1 in the early stages of parasite entry into the host cells has been reported [7]. It is a highly abundant surface protein which is expressed on the rapidly dividing tachyzoites [11]. As the most predominant antigen, it may be used for antibody-based detection [12]. Structural studies showed that SAG1 crystallized as a dimer [8].
This paper describes the primary structure of mature SAG1 of an Indonesian Toxoplasma gondii isolate, sequence comparison with published sequence data of T. gondii strains or isolates and the use of SAG1 in strain determination.
The Indonesian isolate of T. gondii was isolated from the diaphragm of a goat at the slaughter house Cibadak in Sukabumi, West Jawa, Indonesia.
Primers used in PCR reactions allowed the isolation and amplification of mature SAG1 gene. PCR reactions were carried out using PCR beads (Ready-To-Go; Amersham Pharmacia Biotech, USA) in 25 µl buffer, 0.4 µM of each primer, and variable amounts of matrix (genomic DNA or cDNA), at the following conditions: initial denaturation at 94℃ for 2 min; 30 cycles of denaturation at 95℃ for 40 sec, annealing at 60℃ for 40 sec, and elongation at 72℃ for 1 min 30 sec; finally an additional elongation step at 72℃ for 10 min then 4℃. One fifth of the reaction products was analyzed by electrophoresis on a 0.8% agarose gel.
Cloning in pCR2.1-TOPO was carried out using TOPO TA Cloning system (Invitrogen Life Technologies, France), a methodology based on topoisomerase reaction, according to the instruction manual. This system allows direct cloning of PCR reaction products. The topoisomerase reaction mixture contained 1 to 2 µl of the PCR reaction products and 1 µl TOPO vector in 6 µl final volume, and was incubated for 5 min at 22℃. Reaction products were kept on ice or at -20℃ until use.
Transfection was carried out using the TSS method in E. coli DH 5a. TSS-competent E. coli DH 5a bacteria were obtained by concentrating fresh exponential phase bacterial culture DH 5a (OD600nm around 0.6) 10-fold in LB containing 10% PEG 6,000 (w/v), 5% DMSO (v/v) and 35 mM MgCl2. Different amounts of the transfection mix were spread on LB agar plates containing 50 µg/ml ampicilline, 40 µL of 40 mg/ml X-Gal and 40 µl of 100 mM IPTG and incubated at 37℃, for one night (OVN). White bacterial colonies were cultured in 5 ml LB-ampicilline for OVN. Bacteria were harvested by centrifugation (Sorvall, 4,000 rpm, 10-15 min, 4℃). The bacterial pellet was used for plasmid preparation.
Plasmids were prepared using the alkaline lysis method. Briefly, pelleted bacteria were first resuspended in 0.3 ml buffer 1 (50 mM Tris-HCl, pH 7.5, 10 mM EDTA). 0.3 ml buffer 2 (0.2 NaOH, 1% SDS) was then added and the solution mixed without vortexing. Finally 0.3 ml buffer 3 (2.55 M potassium acetate, pH 4.8) was added and the solution mixed without vortexing then centrifuged in a minicencrifuge for 15 min at maximum speed (13,000 rpm) at room temperature. The supernatant (0.8 ml) was then precipitated by the addition of 0.7 ml isopropanol and centrifugation (minicentrifuge, 13,000 rpm, 15 min, room temperature). The pellet was washed with 70% ethanol and slightly dried.
The plasmid pellet was dissolved in 50 µL 10 mM Tris-HCl pH 7.5 and 0.5 mg/ml RNAses and incubated at 37℃ for 30 min. Plasmid analysis was carried out by single digestion with EcoRI or double digestion with BamHI-EcoRI, in 20 µl buffer containing 2.5-5.0 µl plasmid solution, 5 units of each enzyme, at 37℃ for 1 h 30 min. Digestion products were then analyzed by electrophoresis on a 0.8% agarose gel. Visualization was by ethidum bromide and observation under UV lamp.
Sequence alignment was done, and a phylogenetic tree was constructed, using an algorithm established by Corpet [2].
Nucleotide sequencing was performed on the two strands, directly on pGEX-based constructs, using specific primers localized upstream and downstream of the insertion site. The nucleotide sequence obtained and the amino-acid sequence deduced are shown in Fig. 1. Sequencing was done on three independent clones and no divergence was observed between the sequences obtained. The sequence established is therefore the actual sequence of mature SAG1 of the Indonesian Toxoplasma isolate that we have called IS-1 for convenience in this paper (Acc. No. AY651825).
In order to investigate its degree of conservation, and its posible use in strain definition, the sequence of mature SAG1 of the Indonesian isolate was submitted for comparison with sequence data of seven strains or isolates available in GeneBank. Sequence alignment was carried out using an algorithm established by Corpet [2] at the nucleotide (Fig. 2) as well as at the amino-acid level (data not shown). Codon and amino-acid variants are also shown in Table 1.
The overall picture of the results of the sequence comparison between T. gondii strains or isolates showed that in mature SAG1 gene variations affected 15 codons out of 260. At the amino-acid level, they concerned all categories, i.e. uncharged polar amino-acids which are relatively hydrophilic and usually on the outside of the protein surface, non polar ones that have tendency to cluster together on the inside, basic and finally acidic ones.
These last amino-acids, of opposite charge, are very polar and nearly always found on the outside of proteins. By looking more thoroughly at the variations, the first interesting finding was the fact that there were only two possibilities. At the nucleotide level, only two variants, instead of the possible four, were observed. Accordingly, for example at position 97, only codon gtg (V) or gag (E) was used. The two other variations, i.e. gcg (A) and ggg (G) were not found in any of the 8 strains/isolates of T. gondii considered (Table 1). At the codon level, variations were silent (without amino-acid change), conservative (giving rise to amino-acids with the same characteristics) or drastic resulting in an important change of the side chain characteristics, e.g. acidic to basic amino-acid (residue 232) (Table 1). For a given codon, the strains or isolates considered were so divided into two main groups, each belonging to one or the other category.
The frequency of some variations was evenly distributed, reflecting thus actual differences between the two categories, while in others one variant was found in only one strain or isolate (Table 1). In this case the unique variant constituted a unique feature of that strain or isolate and the preferred codons or amino-acids may constitute the original codons or amino-acids in the ancestral organism. According to this point of view, and based on SAG1, the Indonesian isolate IS-1 may be considered as being close to the prototype strain as it possesses the major as well as the preferred variations (Table 1 and see below). Finally, according to its sequence variations, on the whole the IS-1 isolate could be defined as a genuine RH strain as it is 100% homologous to an RH strain, RH2, and only one nucleotide (in codon 96, silent variant) differs from RH1 and CB (also RH strain). More differences were observed between the two RH strains analyzed (RH1 and RH2; codons 96 and 232; Table 1).
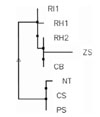
A phylogenetic tree, based on SAG1, was constructed, using an algorithm established by Corpet [2] to determine the parental relationship between 8 Toxoplasma strains or isolates. As they were studied in different parts of the world, they could have divergently evolved depending on the geographical localization. The phylogenetic tree clearly shows that parental relationship appeared to be unrelated to the geographical origins and that the strains or isolates were divided in two major groups (Fig. 3). The Indonesian isolate IS-1 was found in the group that comprised the three RH strains, studied respectively in North America (CB), in Cuba (RH2) and in Europe (RH1) and the Chinese isolate ZS1. ZS1 appeared to be the most distant within the group. The second group included a strain P, a strain C and a strain NT, the last one being studied in China. The phylogenetic tree indicates that the two Chinese isolates were not part of the same group. The phylogenetic tree also shows that the Indonesian isolate undoubtedly belongs to the RH strain. It is not surprising therefore that the RH strain IS-1 isolate is virulent. Sequence comparison clearly showed thus that SAG1 sequence can be used for strain determination.
SAG1 (p30), a highly abundant surface protein which is expressed on the rapidly dividing tachyzoites and which constitutes the most predominant antigen [11], plays a essential role in the early stages of parasite entry into the host cells [7]. Sequence comparison between T. gondii strains or isolates indicated that variations involved all categories of amino acids. Interestingly and curiously, at the nucleotide level only two variants, instead of the possible four, were observed. That limited the extent of mutations as at the very most codon variations lead to only two amino-acid variants. Due to the structural and/or functional constraints, other mutations may lead to non functional SAG1. Owing to the essential role of this protein, those variations are highly detrimental to parasite survival.
The comparison and the phylogenetic tree of SAG1 sequences showed that at the level of SAG1 gene Toxoplasma strains and isolates are divided in two major families, independently of their geographical origin. Isoenzymatic characterization and genetic analyses established that the number of Toxoplasma strains was limited to 2-3 main groups [3,9]. According to Sibley and Boothroyd [13], virulent strains originated from a single lineage which remained genetically homogeneous despite being globally widespread and despite the ability of the organism to reproduce sexually. The limited number of lineages may be explained by an exceedingly rare sexual recombination in natural populations [9]. The Indonesian isolate IS-1 was isolated from the diaphragm of a goat in a slaughter house in West Jawa in 1998. The question is: where did it originate from? It may be a local strain present in Indonesia since a long time ago. By comparing the frequency of the variations in SAG1, it appeared that this isolate is close to the prototype strain as it possesses the major as well as the preferred variations that may be the original constituents in the ancestral organism. Within this context, the RH strain may be considered as being the closest to the ancestral organism. This work undoubtedly showed that SAG1 sequence may be used in Toxoplasma strain determination. The results are in perfect agreement with those obtained by other methodologies. Thanks to its ease of use and its accuracy, the method can be favorably applied to establish the strain of unknown Toxoplasma isolates. We demonstrated in this paper that applied to the Indonesian isolate IS-1, the sequence comparison and the phylogenetic tree of SAG1 gene allowed us to define IS-1 as a genuine virulent RH strain and that the complete homology with RH2 strain is in favour of a possible recent introduction of RH2 strain in Indonesia.
In the primary structure, variations observed in this work were mainly found at the C-terminal half or domain 2 of SAG1 protein and some clustered in restricted areas. None was detected at the N-terminal area or domain 1. Zones between amino acids 174-193 and 221-252 (and particularly between amino-acids 232-234 and 244-252) might constitute hot-spots for variations as they contained the densest variation clusters. Presumably at these positions sequence variations have to result in limited effect on protein function.
Based on the primary structure, a number of antigenic and immunogenic segments of SAG1 have been identified, using two complementary approaches, i.e. determination of antigenic index [1] and use of synthetic peptides in vaccination trials [6]. Except for residues 252 and 264 that are localized in one of the six predicted decapeptides exhibiting the most confidently antigenic index, the sequence variations were observed localized outside of these segments. Residue 252 was also within one of the synthetic peptides. Nevertheless, as recognition by host immune system is also defined by conformational epitopes, we can not exclude the possibility that sequence variations localized outside the predicted linear epitopes may also constitute specific antigenic and immunogenic characters.
The three dimensional structure of Toxoplasma SAG1 has now been established [8]. Information on the structure help to understand how this protein functions. Structural studies showed that SAG1 crystallized as a dimer, each monomer being composed of domain 1 (N-terminal half) and domain 2 (C-terminal half). Owing to the extensive dimer interface and the high strength of monomer-monomer interactions, SAG1 was proposed to also exist as a dimer on the parasite surface [8]. Most of the variations occur at the C-terminal half (domain 2). Three of the amino acid variations are within a β-strand, i.e. variation 97 (V/E) in β-strand g of domain D1 (residues 93-99), variation 126 (L/V) in β-strand b of domain D2 and variation 221 (K/Q) in β-strand g of domain D2. One of the sequence variants was an amino acid involved in hydrogen bonds implicated in monomer interactions, i.e. variation 118 (D/N). It is worthwhile mentioning that at this position five variations were found in the SRS (SAG1-related sequence) superfamily, the amino acids being D, N, E, K, or G [8]. Interestingly, two variants, i.e. residues 186 (K/N) and 174 (T/S), are localized at the protein surface. This might result in epitope variations. None of the variants affect any of the six disulfide bonds involved in the integrity of the three dimensional structure. Finally, variations observed till now do not appear to induce obvious effects on parasite survival, probably because they do not interfere with the structural integrity hence the function of the protein.
Figures and Tables
Fig. 1
Nucleotide and amino-acid sequence of mature SAG1 of the Indonesian Toxoplasma isolate IS-1. Nucleotide sequence (lower row) is presented as codons and the nucleotide numbering is given. The first t of the first codon ttc is taken as nucleotide number 1. The deduced amino-acid sequence (upper row) is presented with one-letter code, and the corresponding amino-acid hence codon numbering indicated.

Fig. 2
Comparison of nucleotide sequence of mature SAG1 gene with SAG1 sequences of 7 other T. gondii strains or isolates. Alignment was carried out using an algorithm developed by Corpet [2]. Sequences are presented as codons. Nucleotide (left, upper) and codon (right, in brackets) numberings are indicated. The stop codon tga (codon no. 260, bases 778-780) is underlined and in italics. Codons with nucleotide variation are underlined and the variations indicated by bold characters. Con, consensus sequence. Sequence accession numbers are as follows: IS-1, AY651825 (Indonesian isolate); CB, X14080 (clone P30.5Cos1); CS, S63900 (strain C); RH1, AY217784 (strain RH); RH2, S76248 (strain RH); PS, S85174 (strain P); ZS, S73634 (Chineese isolate); NT, AF110182 (strain NT).

References
1. Alix AJP. Predictive estimation of protein linear epitopes by using the program PEOPLE. Vaccine. 1999. 18:311–314.

2. Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988. 16:10881–10890.

3. Cristina N, Dard ML, Boudin C, Tavernier G, Pestre-Alexandre M, Ambroise-Thomas P. A DNA fingerprinting method for individual characterization of Toxoplasma gondii strains: combination with isoenzymatic characters for determination of linkage groups. Parasitol Res. 1995. 81:32–37.

4. Dobrowolski JM, Sibley LD. Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell. 1996. 84:933–939.

5. Dubremetz JF. Host cell invasion by Toxoplasma gondii. Trends Microbiol. 1998. 6:27–30.
6. Godard I, Estaquier J, Zenner L, Bossus M, Auriault C, Darcy F, Gras-Masse H, Capron A. Antigenicity and immunogenicity of P30-derived peptides in experimental models of toxoplasmosis. Mol Immunol. 1994. 31:1353–1363.

7. Grimwood J, Smith JE. Toxoplasma gondii: the role of parasite surface and secreted proteins in host cell invasion. Int J Parasitol. 1996. 26:169–173.

8. He XL, Grigg ME, Boothroyd JC, Garcia KC. Structure of the immunodominant surface antigen from the Toxoplasma gondii SRS superfamily. Nat Struct Biol. 2002. 9:606–611.
9. Howe DK, Sibley LD. Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J Infect Dis. 1995. 172:1561–1566.

10. Jung C, Lee CY, Grigg ME. The SRS superfamily of Toxoplasma surface proteins. Int J Parasitol. 2004. 34:285–296.
11. Kasper LH, Bradley MS, Pfefferkorn ER. Identification of stage-specific sporozoite antigens of Toxoplasma gondii by monoclonal antibodies. J Immunol. 1984. 132:443–449.
12. Kimbita EN, Xuan X, Huang X, Miyazawa T, Fukumoto S, Mishima M, Suzuki H, Sugimoto C, Nagasawa H, Fujisaki K, Suzuki N, Mikami T, Igarashi I. Serodiagnosis of Toxoplasma gondii infection in cats by enzyme-linked immunosorbent assay using recombinant SAG1. Vet Parasitol. 2001. 102:35–44.

13. Sibley LD, Boothroyd JC. Virulent strains of Toxoplasma gondii comprise a single clonal lineage. Nature. 1992. 359:82–85.

14. Hartati S, Wuryastuti H, Widada S, Sumartono , Kusumawati A. Kloning dan ekspresi gen penyandi SAGI Toxoplasma gondii isolat lokal pada vektor pGEX-2T. J Sain Vet. 2004. 22:19–22.
15. Hartati S, Widada S, Sumartono , Kusumawati A. Cloning of the gene encoding SAGI of local isolate Toxoplasma gondii in E. coli DH5á. J Sain Vet. 2003. 21:51–56.
17. Zangerle R, Allerberger F, Pohl P, Fritsch P, Dierich MP. High risk of developing toxoplasmic encephalitis in AIDS patients seropositive to Toxoplasma gondii. Med Microbiol Immunol. 1991. 180:59–66.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download