Abstract
Outbreaks of porcine epidemic diarrhea (PED) have resulted in significant economic losses in the swine industry, and another PED outbreak occurred in 2014 in Korea. Isolating and culturing PED virus (PEDV) allow investigations into its pathogenesis and the development of vaccines and diagnostic assays. In this study, we successfully isolated two PEDV isolates (QIAP1401 and QIAP1402) from naturally infected piglets at Jeju-do, Korea. Viral propagation was confirmed in Vero cells based on cytopathic effect, immunofluorescence assay, reverse transcription-polymerase chain reaction, and electron microscopic analyses. The QIAP401 isolate propagated well in Vero cells for 70 passages, with titers of 106.5 to 107.0 50% tissue culture infectious dose/mL, which increased gradually with passaging. The nucleotide and amino acid sequences of the QIAP1401 isolate were determined and compared with those of other PEDV isolates. The QIAP1401 isolate was determined to be closely related to the USA/Minnesota271/2014 strain (> 99.9% nucleotide similarity) that was isolated in the USA in 2014. Phylogenetic analysis based on several PEDV genes suggested that a new PEDV variant is circulating in the Korean swine industry, with 93.08% similarity to the SM98 strain isolated in 1998. In addition, the QIAP1401 strain showed strong virulence in 3-day-old piglets and 11-week-old growing pigs.
Porcine epidemic diarrhea virus (PEDV) induces vomiting, severe watery diarrhea, and dehydration in neonatal piglets, eventually leading to death. Since PEDV was first isolated (CV777 strain) in Belgium in 1978 [19], PED outbreaks have been identified regularly in Europe and Asia, including Korea, Japan, China, Taiwan, and Vietnam [71020]. More recently, a PED outbreak occurred suddenly in the USA in 2013 and spread to Canada and Mexico [117]. Since a live-attenuated PEDV vaccine was developed from a Korean PEDV isolate (SM98 strain) obtained from naturally infected piglets in Korea in 1998, PED vaccines have been used in pig farms to prevent PED outbreaks [11]. However, PED continues to be a major threat to the Korean pork industry [21]. In March 2014, a large number of PEDV infections occurred unexpectedly in many Korean pig farms, which had a significant economic impact on swine farms due to a high mortality rate among newborn piglets [12].
Similar to transmissible gastroenteritis virus, the PEDV belongs to the family Coronaviridae, subfamily Coronavirinae, genus Alphacoronavirus and contains a single-stranded positive-sense RNA genome of approximately 28 kb [20]. The PEDV genome is composed of at least seven open reading frames (ORFs), encoding three non-structural proteins (ORF1a, 1b, and ORF3) and four structural proteins; spike (S), envelope, membrane (M), and nucleocapsid (N) proteins. The S glycoprotein mediates neutralizing antibodies in pigs and viral attachment to target cells and has been reported to bind to carbohydrate (sialic acid) and proteinaceous (aminopeptidase N) cell surface molecules [14]. The M protein has an important role in assembly of the viral nucleocapsid and membrane [23]. The N protein binds to viral RNA and provides a structural basis for the helical nucleocapsid [6]. In addition, the N protein participates in transcription of the viral genome, formation of the viral core, and packaging of viral RNA [20]. Therefore, investigation of structural proteins is suitable for elucidating the epidemiological and molecular statuses of PEDV isolates.
The advent of new mutant PEDV isolates will allow study of PED pathogenesis, effects of disinfectants, and the development of antigenic or serological diagnostic assays and a live-attenuated or inactivated vaccine to prevent PED outbreaks [2]. Growing pure PEDV in a cell culture system has been attempted in many veterinary laboratories but has proven difficult because of many factors, such as fecal toxins, other viral contaminants, and the trypsin concentration. Although PEDV can be detected in cells after one or two passages, the virus tends to lose infectivity after additional passages. A few reports are available regarding successful culture of Korean PEDV isolates since the first isolation of PEDV [111221]. In this study, we attempted to isolate field PEDV from 30 PEDV-positive samples by using Vero cells. Two PEDV strains, designated QIAP1401 and QIAP1402, were isolated successfully, and the QIAP1401 strain was passaged 70 times in cell culture. We characterized the biological properties of the QIAP1401 strain during serial passage, and the nucleotide sequences of the QIAP1401 strain structural genes were determined after specific passages to evaluate genetic relationships. In addition, we investigated the pathogenicity of the QIAP1401 strain in 3-day-old piglets and 11-week-old pigs.
Animal ethic committee of Animal and Plant Quarantine Agency (APQA; Gimcheon, Korea) approved the animal experimental design (approval No. 2015-283).
Thirty small intestinal homogenates from pigs diagnosed with PED based on clinical signs were selected for virus isolation at the Jeju and Gangwon-do Veterinary Service Laboratory. Small intestinal tissues were used to generate a 10% homogenate in Dulbecco's modified Eagle's medium (DMEM) containing 2.0 µg/mL crystalized trypsin (Sigma-Aldrich, USA). After preparing the 10% homogenate, the samples were centrifuged at 4,000 × g for 15 min at 4℃. The supernatant was filtered through a 0.45 µm membrane (Millipore, USA) and used for virus isolation.
The Vero cells (ATCC CCL-81 and ATCC CCL-1586) used for virus isolation were cultured in DMEM supplemented with antibiotics (100 IU/mL penicillin, 10 µg/mL streptomycin, and 0.25 µg/mL amphotericin B) and 10% heat-inactivated fetal bovine serum (Gibco BRL, USA). The cells were cultured in 24-well plates at 37℃ in a 5% CO2 incubator. Confluent Vero cells were washed twice with phosphate buffered saline (PBS, pH 7.2) and inoculated with 100 µL of the final samples for 1 h at 37℃. After adding 200 µL DMEM, the 24-well plates were incubated for 2 h at 37℃ in a CO2 incubator. After incubation, the samples were discarded, and 1 mL fresh DMEM containing 1 µg/mL crystalized trypsin was added to each well. The plates were incubated at 37℃ in 5% CO2 for 5 days. When cytopathic effects (CPEs) were detected in the cells, the supernatant was harvested for further passage and inoculated into newly prepared Vero cells for the second passage. If no CPEs were observed 5 days after harvesting the supernatant, the Vero cells were fixed in 80% cold acetone and subjected to an immunofluorescence assay (IFA). If CPEs were not detected, and the IFA results were negative after three passages, the sample was considered negative for the virus.
Viral titration was conducted in 96-well microplates by using 10-fold serial dilutions, and CPEs were observed under a microscope at 5 days post-inoculation (DPI). The viral titers were calculated according to the Reed and Muench method and expressed as the 50% tissue culture infectious dose (TCID50/mL).
Vero cells were plated and infected with the QIAP1401 strain in 24- or 96-well plates and fixed in cold acetone (−20℃) for 20 min. After three successive washes with PBS (pH 7.2), the cells were reacted with a 7G7 mouse monoclonal antibody (APQA, Korea) specific to PEDV for 45 min at 37℃ and then stained with a 100× dilution of fluorescein isothiocyanate-conjugated goat-anti-mouse IgG + IgM (KPL Laboratories, USA). After washing the plates with PBS, the Vero cells were air-dried and examined at 200× under a fluorescence microscope (Nikon, Japan). Samples showing specific fluorescence in the cytoplasm were considered positive.
Viral RNA extraction from 200 µL of the small intestinal homogenate samples was performed by using a viral RNA isolation kit (iNtRON Biotechnology, Korea). Viral RNA was eluted using 50 µL of elution buffer, and the eluted RNA was subjected to conventional reverse transcription-polymerase chain reaction (RT-PCR) using the i-TGEV/PEDV detection kit (iNtRON Biotechnology, Korea) and following the manufacturer's instructions. The PCR products were visualized via 1.8% agarose gel electrophoresis and ethidium bromide staining. Samples showing a 525 bp band were considered positive.
A commercial rapid detection kit for detecting PEDV was applied to 30 intestinal homogenate samples and two PED isolates according to the manufacturer's instructions (Bionotes, Korea). Briefly, a 10% suspension of each intestinal sample was prepared, and a 100 µL aliquot of the sample was applied to the sample well. The results were read 5 min after applying the sample, and the appearance of two lines (test line and control line) was considered positive. The formation of a single line in the control area was considered a negative result.
Vero cells infected with the QIAP1401 isolate were harvested 36 h post-inoculation and were frozen and thawed three times. After centrifugation at 4,000 × g for 30 min to remove cell debris, the viral supernatant was treated with 10% polyethylene glycol (MW 8000; Sigma-Aldrich, USA) and 0.5 M NaCl and precipitated overnight at 4℃. After precipitation, the pellet was resuspended in GTNT buffer (20 mM glycine, 100 mM Tris-Cl, 100 mM NaCl, and 1 mM ethylenediaminetetraacetic acid; pH 7.6) at 1% of the original volume. The suspension was layered on top of a 20% to 50% sucrose solution and centrifuged at 100,000 × g for 3 h in a SW-41 rotor (Beckman, USA). The band between the 20% and 30% sucrose layers was collected and dialyzed with PBS solution to eliminate the residual sucrose overnight at 4℃. One drop of the purified PEDV was placed on Formvar-coated grids and stained negatively with 1% uranyl acetate. The PEDV particles were examined under an electron microscopy (EM) (Hitachi 7100; Hitachi, Japan).
Viral RNA was extracted from the QIAP1401 and QIAP1402 isolates by using an RNA extraction kit (Qiagen, Germany) according to the manufacturer's instructions. The extracted RNA was eluted in 50 µL RNase- and DNase-free water. RT-PCR was carried out with specific primers to amplify the ORF3, S, M, and N regions of PEDV (Table 1). The RT-PCR was performed in a reaction mixture containing 10 µL denatured RNA, 1 µL each primer (50 pmol), 10 µL 5× buffer (12.5 mM MgCl2), 2 µL dNTP mix, 2 µL enzyme mix (reverse transcriptase and Taq polymerase), and 24 µL distilled water (Qiagen). The cycling profile consisted of cDNA synthesis at 42℃ for 30 min, followed by 35 cycles of 95℃ for 45 sec, 50℃ for 45 sec, and 72℃ for 1 min, with a final extension at 72℃ for 5 min. The PCR products were visualized via 1.8% agarose gel electrophoresis and ethidium bromide staining. All PCR products purified using the gel extraction kit were ligated into the pGEM-T Easy vector system (Promega, USA) according to the manufacturer's protocol. Plasmid DNA was isolated from amplified Escherichia coli (DH5α), and recombinant plasmids were identified by EcoRI enzyme digestion (Bioneer, Korea). The sequences of the purified plasmids were analyzed by using the MJ Research PTC-225 Peltier Thermal Cycler and ABI PRISM BigDye Terminator Cycle Sequencing kits with AmpliTaq DNA polymerase (Applied Biosystems, USA), according to the manufacturers' protocols. Single-pass sequencing was performed for each template by using universal primers (e.g., SP6 and T7). The fluorescently labeled fragments were purified from the unincorporated terminators by applying an ethanol precipitation protocol. The samples were resuspended in distilled water and subjected to electrophoresis using the ABI 3730xl sequencer (Applied Biosystems). Both DNA strands were sequenced to verify the sequences.
Phylogenetic analysis of the nucleotide sequences from the two PEDV isolates and 25 other PEDV strains obtained from the GenBank database (National Center for Biotechnology Information [NCBI], USA) was performed. Multiple sequence alignments and sequence similarity calculations were performed by using DNASTAR (DNASTAR, USA) and DNASIS software (Hitachi Solution, USA), respectively. Phylogenic analyses were conducted by applying the neighbor-joining method using MEGA version 7.0.18 software [9]. The bootstrap method with 1,000 replicates was applied to determine the reliability of the inferred phylogenetic tree.
Group 1 consisted of four 3-day-old piglets, previously non-vaccinated for PEDV, purchased from a commercial pig farm that was PEDV- and maternal antibody-free. All pigs were housed at 32℃ in a pig isolator. Two pigs were orally administered a 1 mL dose of 103.0 TCID50/mL QIAP1401-P70 via 5 mL pipette, and the remaining two pigs received no treatment. Group 2 comprised seven 11-week-old growing pigs seronegative for PEDV. The pigs were orally administered 1 mL 105.0 TCID50/mL QIAP1401-P70 via 5 mL pipette. All pigs were monitored daily for clinical signs of PEDV infection, such as watery diarrhea and dehydration. Clinical or RT-PCR scores were divided into four grades: 0, 1, 2, and 3. Stool samples were collected daily from all pigs using cotton-tipped swabs and were subjected to the rapid detection and RT-PCR kits to detect the PEDV antigen. The four piglets in group 1 were necropsied. Small intestinal tissues collected from each pig were fixed in 10% formalin and embedded in paraffin according to standard procedures. The deparaffinized intestinal tissue sections were stained with hematoxylin and eosin for histopathologic examination. A PEDV-N specific monoclonal antibody was used for immunohistochemistry (IHC) assay. The antigen was detected by using an avidin-biotin kit (Ventana Medical System, USA) and an automatic immunohistochemical stainer (Ventana Medical System). Epithelial cells within intestinal samples that turned brown were considered positive. The pigs in group 2 were observed for 3 weeks post-inoculation, but were not necropsied.
Among 30 intestinal homogenate samples, 26 and 22 tested positive in the RT-PCR and rapid detection kit assessments, respectively. Virus isolation was attempted using the 30 intestinal homogenates in Vero cells. Two PEDV isolates, designated QIAP1401 and QIAP1402, grew successfully in Vero cells inoculated with small intestinal samples from piglets from the Jeju-do pig farms (collected 26 May 2014). As shown in Fig. 1, clear CPEs, characterized by cell fusion and large syncytia, were observed in the QIAP1401 and QIAP1402 isolates after the second passage. The supernatants from the two isolates were analyzed by the rapid detection kit and were positive for PEDV presence. No difference in CPEs was observed between the QIAP1401 and QIAP1402 isolates. The two PEDV isolates were further passaged serially to confirm efficient propagation. Distinct CPEs were usually observed in Vero cells infected with each isolate within 30 h post-inoculation, and large cells containing many nuclei and floating cells were observed over time. After fixing the cells with cold acetone and staining with the 7G7 monoclonal antibody, specific fluorescence was detected in the cytoplasm of Vero cells infected with QIAP1401 and QIAP1402 (Fig. 2). The QIAP1401 strain was serially passaged 70 times in Vero cells, and the viral titers of QIAP1401-P10, -P40, and -P70 cultured in roller bottles were 106.5, 106.8, and 107.0 TCID50/mL, respectively (panel A in Fig. 2). The QIAP1401-P10, -P40 and -P70 viral titers peaked at 105.7, 106.0, and 106.7 TCID50/mL, respectively, at 36 h post-inoculation (panel B in Fig. 2). We concluded that the QIAP1401 strain adapted to the Vero cells because the titer increased continuously until passage 70.
PEDV particles from the QIAP1401 strain purified by gradient sucrose density centrifugation were examined by EM. As shown in Fig. 3, the viral particles were 80 to 120 nm in diameter with crown-shaped projections, indicating a morphological appearance typical of Coronaviridae.
A 4161 bp sequence of the S gene from the two PEDV isolates was identified, and the amino acid sequence was deduced. The nucleotide sequence of the QIAP1401 strain was deposited in GenBank (NCBI) under accession number KX793713. The nucleotide sequences from the two Korean PEDV isolates were compared with those from 25 PEDV strains retrieved from GenBank to analyze the epidemiological relationships among PEDVs. A phylogenetic tree based on the nucleotide sequences of the S gene revealed that the Korean PEDV strains isolated in 2014 belong to the G2 group, with the QIAP1401 and QIAP1402 strains most closely related to the USA strain designated USA/Minnesota271/2014 (Fig. 4). The similarities of the PEDV S gene nucleotide sequences were 99.6% to 99.7% among the Korean isolates obtained after 2013 and 93.0% to 96.6% among QIAP1401 and the Korean PEDV isolates obtained before 2011. Alignment of the deduced amino acid sequences showed 99.4% to 99.7% similarity among QIAP1401 and the Korean PEDV strains obtained after 2013.
The complete S, ORF3, M, and N genes were sequenced after passages 3, 20, 40, and 60, respectively. As shown in Table 2, QIAP1401 after 60 passages revealed 12 amino acid changes compared with the original QIAP1401 strain. Among the four structural proteins, the S protein had the highest mutation rate of 58.3% (7/12). No deletions were detected in the four QIAP1401 structural proteins after 60 passages (data not shown).
Two piglets were orally administered the QIAP1401-P70 strain, and another two piglets were housed together with the inoculated piglets in a pig isolator for direct contact exposure. All piglets started to exhibit clinical PED signs, such as vomiting and diarrhea, at 1 DPI and proceeded to have severe watery diarrhea at 2 DPI (panel A in Fig. 5). One of the control piglets died of dehydration at 4 DPI, and the remaining three piglets died of dehydration at 5 and 6 DPI. All feces obtained from the four piglets at 1 to 6 DPI were positive for PEDV as determined by both RT-PCR and the rapid detection kit (panel A in Fig. 5). The dead piglets were necropsied on the day they died and showed shortened and fused small intestinal villi on microscopic examination (panel A in Fig. 6). In addition, the PEDV antigen was detected in the jejunal epithelial cells of atrophied villi by IHC (panel B in Fig. 6).
Seven growing, 11-week-old, PEDV-seronegative pigs were administered the same PEDV strain and housed in a compartment. As shown in Fig. 5B, the clinical signs began with mild diarrhea at 2 DPI. The pigs exhibited the most severe clinical signs at 4 DPI and returned to normal with no clinical symptoms at 11 DPI. The PEDV antigen was first detected in feces by RT-PCR at 1 DPI, and the greatest antigen level was detected at 3 and 4 DPI. The PEDV antigen in feces was not detected at 11 DPI in growing pigs, all of whom survived administration of QIAP1401-P70.
PED has been reported in many countries including Korea, and PEDV, which causes severe diarrhea in piglets, has led to enormous economic losses in the Korean pig industry. A total of 669 cases of the disease occurred between 2000 and 2015, and two large epidemics were observed in 2003 and 2014 (Fig. 7). In 2014, 169 PED outbreaks causing high mortality were identified, officially, and more than 33,600 piglets were infected with PEDV at 169 Korean pig farms.
As several PEDV cases were diagnosed in 2014, attempts were made to isolate PEDV from infected pigs or feces. As a result, several Korean isolates, such as the KPV1405, KNU1307, and BM1 strains isolated from the feces of naturally infected piglets, were reported [412]. We attempted to isolate PEDV from 30 intestinal homogenates and obtained the QIAP1401 and QIP1402 strains from Vero cells. They were confirmed to be PEDV based on CPEs, IFA, EM, and RT-PCR results. An appropriate trypsin concentration and Vero cell line were essential for isolating PEDV in cell culture [813]. In this study, we found that ATCC-CCL81 Vero cells were more suitable for propagating field PEDV than were ATCC-1586 Vero cells, and 2 µg/mL trypsin at 37℃ for 1 h before inoculation into the Vero cells and medium containing 1 or 1.5 µg/mL crystalized trypsin were critical for propagating PEDV (data not shown). Subsequently, one of the new PEDV isolates has been used to develop a vaccine candidate for live and inactivated PEDV vaccines. New vaccines based on the current strains are needed because the PEDV vaccine based on the SM98 or DR13 strains might be less related antigenically to the re-emerged PEDV strains [241822].One of the two isolates in this study was serially propagated 70 times in Vero cells. The viral titer of QIAP1401 reached 107.0 TCID50/mL by passage 70, indicating that the QIAP1401-P70 strain was adapted to the Vero cells and could be used as a vaccine strain.
Nucleotide sequence and phylogenetic analyses based on the S gene of 27 PEDV strains revealed that the QIAP1401 and QIAP1402 strains are almost identical to the USA PEDV strains isolated in 2014. These strains were confirmed to be variants of the PEDV strain belonging to the G2 group, indicating that the QIAP1401 strain is a variant of the strains used in traditional vaccines such as SM98 or DR13. If this relationship is factual, it remains unclear how the PEDV variant was introduced into Korea. A molecular epidemiologist suggested that the Korean PEDV variant strains may have been transmitted directly or indirectly from USA PEDV strains that circulated in 2013 and 2014 [3]. The SM98-1 strain has a 52-nucleotide deletion between the end of the S protein and the start of ORF3 [12]. The QIAP1401 strain was passaged 70 times in Vero cells and showed 11 amino acid changes in the four structural proteins after passage 20 compared with those of the original QIAP1401 strain; these 11 mutated amino acids persisted for 60 passages. Only one amino acid change in the M protein was identified at passage 60, indicating that the QIAP1401 strain is genetically stable over serial passages in Vero cells.
The pathogenicity of the PEDV strains has been investigated extensively in pigs of different ages, and most of the PEDV strains isolated from naturally infected pigs are strongly virulent [21524]. In this study, PEDV-seronegative 3-day-old piglets and 11-week-old growing pigs were administered the QIAP1401-P70 strain orally to investigate the pathogenicity of the QIAP1401 strain. All four of the 3-day-old piglets, including two non-inoculated piglets, shed the virus in their feces, developed severe watery diarrhea from 1 to 6 DPI, and died at 4, 5 or 6 DPI, indicating that the QIAP1401-P70 strain is highly pathogenic to newborn piglets.
PEDV is infectious in pigs at all stages, and it produces mild diarrhea in adults and severe diarrhea in newborn piglets, with up to a 100% mortality rate [520]. Madson et al. [16] reported that 3- and 4-week-old pigs infected with a US PEDV isolate resulted in mild to moderate clinical disease. In this study, 11-week-old growing pigs inoculated with the QIAP1401-P70 strain were positive 1 to 10 DPI by RT-PCR and positive 2 to 10 DPI based on clinical scores. However, we could not detect virus shed in feces by RT-PCR or by the clinical score at ≥ 11 DPI, indicating that the QIAP1401 strain induces moderate to severe watery diarrhea during a relatively short period in 11-week-old pigs.
In conclusion, we isolated two PEDV isolates from naturally infected piglets and confirmed them to be PEDV variants closely related genetically to a USA PEDV stain. We adapted the QIAP1401 strain to Vero cell culture after serial passaging. In addition, the QIAP1401 strain, which is strongly pathogenic to 3-day-old piglets, could be used as a challenge strain to examine the efficacy of inactivated or live-attenuated PEDV vaccines.
Figures and Tables
Fig. 1
Cytopathic effects of (A–C), and immunofluorescence results for (D–F), porcine epidemic diarrhea virus isolates in infected Vero cells at 200×. Vero cells were infected with the QIAP1401 or QIAP1402 isolates and maintained in DMEM containing 1 µg/mL trypsin
Fig. 2
Virus titers according to number of the porcine epidemic diarrhea virus (PEDV) passages (A) and growth curves of the QIAP1401-P10, -P40, and -P70 strains according to time of harvesting in Vero cells (B). TCID, tissue culture infectious dose.

Fig. 3
Virus particles from the QIAP1401 strain propagated in Vero cells. Negatively stained porcine epidemic diarrhea virus particles of 80 to 100 nm in diameter are visible (100,000×). Scale bar = 100 nm.

Fig. 4
Phylogenetic analysis based on the nucleotide sequences of the spike (S) gene of porcine epidemic diarrhea viral strains. The QIAP1401 and QIAP1402 strains were predicted to belong to porcine epidemic diarrhea virus group G2. The phylogenetic tree was constructed based on aligned nucleotide sequences obtained by using the neighbor-joining method. KOR, Korea; CAN, Canada; JPN, Japan; CHN, China; BEL, Belgium; G1, genotype 1; G2a, genotype 2a; G2b, genotype 2b.

Fig. 5
Mean clinical symptom scores for 3-day-old piglets (A) and 11-week-old growing pigs (B) infected with the QIAP1401-P70 strain, and the mean score obtained by reverse transcription-polymerase chain reaction in feces following oral administration of porcine epidemic diarrhea viral strains. RT-PCR, reverse transcription-polymerase chain reaction.
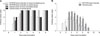
Fig. 6
Jejunal sections of piglets infected with the QIAP1401 strain. Severe atrophy of villi and several cytoplasmic vacuoles were observed in intestinal epithelial cells following hematoxylin and eosin staining (20×) (A); brown staining, indicating a positive reaction, was identified by immunohistochemistry (20×) (B).

Fig. 7
Number of porcine epidemic diarrhea (PED) cases ( ) and PED virus-infected piglets (◆) in Korea between January 2000 and December 2016.
) and PED virus-infected piglets (◆) in Korea between January 2000 and December 2016.

Acknowledgments
We thank the veterinarians who collected the infected piglets in Jeju-do. This study was supported financially by a grant (B-1543083-2015-17-01) from the Animal and Plant Quarantine Agency, Ministry of Agriculture, Food and Rural Affairs (MAFRA), Republic of Korea.
References
1. Chen Q, Gauger PC, Stafne MR, Thomas JT, Madson DM, Huang H, Zheng Y, Li G, Zhang J. Pathogenesis comparison between the United States porcine epidemic diarrhoea virus prototype and S-INDEL-variant strains in conventional neonatal piglets. J Gen Virol. 2016; 97:1107–1121.

2. Chen Q, Li G, Stasko J, Thomas JT, Stensland WR, Pillatzki AE, Gauger PC, Schwartz KJ, Madson D, Yoon KJ, Stevenson GW, Burrough ER, Harmon KM, Main RG, Zhang J. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J Clin Microbiol. 2014; 52:234–243.

3. Choi JC, Lee KK, Pi JH, Park SY, Song CS, Choi IS, Lee JB, Lee DH, Lee SW. Comparative genome analysis and molecular epidemiology of the reemerging porcine epidemic diarrhea virus strains isolated in Korea. Infect Genet Evol. 2014; 26:348–351.

4. Chung HC, Nguyen VG, Moon HJ, Lee JH, Park SJ, Lee GE, Kim HK, Noh YS, Lee CH, Goede D, Park BK. Isolation of porcine epidemic diarrhea virus during outbreaks in South Korea, 2013-2014. Emerg Infect Dis. 2015; 21:2238–2240.

5. Crawford K, Lager K, Miller L, Opriessnig T, Gerber P, Hesse R. Evaluation of porcine epidemic diarrhea virus transmission and the immune response in growing pigs. Vet Res. 2015; 46:49.

6. Duarte M, Tobler K, Bridgen A, Rasschaert D, Ackermann M, Laude H. Sequence analysis of the porcine epidemic diarrhea virus genome between the nucleocapsid and spike protein genes reveals a polymorphic ORF. Virology. 1994; 198:466–476.

7. Fan H, Zhang J, Ye Y, Tong T, Xie K, Liao M. Complete genome sequence of a novel porcine epidemic diarrhea virus in south China. J Virol. 2012; 86:10248–10249.

8. Hofmann M, Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J Clin Microbiol. 1988; 26:2235–2239.

9. Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; 33:1870–1874.

10. Kusanagi K, Kuwahara H, Katoh T, Nunoya T, Ishikawa Y, Samejima T, Tajima M. Isolation and serial propagation of porcine epidemic diarrhea virus in cell cultures and partial characterization of the isolate. J Vet Med Sci. 1992; 54:313–318.

11. Kweon CH, Kwon BJ, Jung TS, Kee YJ, Hur DR, Hwang EK. Isolation of porcine epidemic diarrhea virus (PEDV) in Korea. Korean J Vet Res. 1993; 33:249–254.
12. Lee S, Kim Y, Lee C. Isolation and characterization of a Korean porcine epidemic diarrhea virus strain KNU-141112. Virus Res. 2015; 208:215–224.

13. Li RF, Tian XQ, Liu Y, Xu J, Liu DY. Isolation and genetic analysis of a variant porcine epidemic diarrhea virus in China. Pol J Vet Sci. 2016; 19:65–73.

14. Li W, van Kuppeveld FJM, He Q, Rottier PJM, Bosch BJ. Cellular entry of the porcine epidemic diarrhea virus. Virus Res. 2016; 226:117–127.

15. Lin CM, Saif LJ, Marthaler D, Wang Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016; 226:20–39.

16. Madson DM, Magstadt DR, Arruda PH, Hoang H, Sun D, Bower LP, Bhandari M, Burrough ER, Gauger PC, Pillatzki AE, Stevenson GW, Wilberts BL, Brodie J, Harmon KM, Wang C, Main RG, Zhang J, Yoon KJ. Pathogenesis of porcine epidemic diarrhea virus isolate (US/Iowa/18984/2013) in 3-week-old weaned pigs. Vet Microbiol. 2014; 174:60–68.

17. Ojkic D, Hazlett M, Fairles J, Marom A, Slavic D, Maxie G, Alexandersen S, Pasick J, Alsop J, Burlatschenko S. The first case of porcine epidemic diarrhea in Canada. Can Vet J. 2015; 56:149–152.
18. Park S, Kim S, Song D, Park B. Novel porcine epidemic diarrhea virus variant with large genomic deletion, South Korea. Emerg Infect Dis. 2014; 20:2089–2092.

19. Pensaert MB, de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol. 1978; 58:243–247.

20. Saif LJ, Pensaert MB, Sestack K, Yeo SG, Jung K. Coronaviruses. In : Straw BE, Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, editors. Diseases of Swine. Ames: Wiley-Blackwell;2012. p. 501–524.
21. Song D, Moon H, Kang B. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin Exp Vaccine Res. 2015; 4:166–176.

22. Song D, Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012; 44:167–175.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download