Abstract
The aim of this study was to investigate the molecular characteristics and to conduct a comparative genomic analysis of Mycobacterium (M.) bovis strain 1595 isolated from a native Korean cow. Molecular typing showed that M. bovis 1595 has spoligotype SB0140 with mycobacterial interspersed repetitive units– variable number of tandem repeats typing of 4-2-5-3-2-7-5-5-4-3-4-3-4-3, representing the most common type of M. bovis in Korea. The complete genome sequence of strain 1595 was determined by single-molecule real-time technology, which showed a genome of 4351712 bp in size with a 65.64% G + C content and 4358 protein-coding genes. Comparative genomic analysis with the genomes of Mycobacterium tuberculosis complex strains revealed that all genomes are similar in size and G + C content. Phylogenetic analysis revealed all strains were within a 0.1% average nucleotide identity value, and MUMmer analysis illustrated that all genomes showed positive collinearity with strain 1595. A sequence comparison based on BLASTP analysis showed that M. bovis AF2122/97 was the strain with the greatest number of completely matched proteins to M. bovis 1595. This genome sequence analysis will serve as a valuable reference for improving understanding of the virulence and epidemiologic traits among M. bovis isolates in Korea.
References
1. Achtman M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol. 2008; 62:53–70.

2. Allix C, Walravens K, Saegerman C, Godfroid J, Supply P, Fauville-Dufaux M. Evaluation of the epidemiological relevance of variable-number tandem-repeat genotyping of Mycobacterium bovis and comparison of the method with IS6110 restriction fragment length polymorphism analysis and spoligotyping. J Clin Microbiol. 2006; 44:1951–1962.
3. Bailo R, Bhatt A, Aínsa JA. Lipid transport in Mycobacterium tuberculosis and its implications in virulence and drug development. Biochem Pharmacol. 2015; 96:159–167.
4. Belisle JT, Mahaffey SB, Hill PJ. Isolation of Mycobacterium species genomic DNA. Methods Mol Biol. 2009; 465:1–12.
5. Bouklata N, Supply P, Jaouhari S, Charof R, Seghrouchni F, Sadki K, El Achhab Y, Nejjari C, Filali-Maltouf A, Lahlou O, El Aouad R. Molecular typing of Mycobacterium tuberculosis complex by 24-locus based MIRU-VNTR typing in conjunction with spoligotyping to assess genetic diversity of strains circulating in Morocco. PLoS One. 2015; 10:e0135695.
6. Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, Parkhill J, Rajandream MA. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008; 24:2672–2676.

7. Cho YJ, Yi H, Chun J, Cho SN, Daley CL, Koh WJ, Shin SJ. The genome sequence of ‘Mycobacterium massiliense' strain CIP 108297 suggests the independent taxonomic status of the Mycobacterium abscessus complex at the subspecies level. PLoS One. 2013; 8:e81560.
8. Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999; 27:4636–4641.

9. Forrellad MA, Klepp LI, Gioffré A, Sabio y García J, Morbidoni HR, de la Paz Santangelo M, Cataldi AA, Bigi F. Virulence factors of the Mycobacterium tuberculosis complex. Virulence. 2013; 4:3–66.
10. Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007; 57:81–91.

11. Hotter GS, Collins DM. Mycobacterium bovis lipids: virulence and vaccines. Vet Microbiol. 2011; 151:91–98.
12. Humblet MF, Boschiroli ML, Saegerman C. Classification of worldwide bovine tuberculosis risk factors in cattle: a stratified approach. Vet Res. 2009; 40:50.

13. Jackson M, Stadthagen G, Gicquel B. Long-chain multiple methyl-branched fatty acid-containing lipids of Mycobacterium tuberculosis: biosynthesis, transport, regulation and biological activities. Tuberculosis (Edinb). 2007; 87:78–86.
14. Je S, Ku BK, Jeon BY, Kim JM, Jung SC, Cho SN. Extent of Mycobacterium bovis transmission among animals of dairy and beef cattle and deer farms in South Korea determined by variable-number tandem repeats typing. Vet Microbiol. 2015; 176:274–281.
15. Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008; 36:D250–254.

16. Jeon B, Je S, Park J, Kim Y, Lee EG, Lee H, Seo S, Cho SN. Variable number tandem repeat analysis of Mycobacterium bovis isolates from Gyeonggi-do, Korea. J Vet Sci. 2008; 9:145–153.
17. Jeon BY, Kim SC, Je S, Kwak J, Cho JE, Woo JT, Seo S, Shim HS, Park BO, Lee SS, Cho SN. Evaluation of enzyme-linked immunosorbent assay using milk samples as a potential screening test of bovine tuberculosis of dairy cows in Korea. Res Vet Sci. 2010; 88:390–393.

18. Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997; 35:907–914.
19. Käser M, Ruf MT, Hauser J, Pluschke G. Optimized DNA preparation from mycobacteria. Cold Spring Harb Protoc. 2010; 2010:pdb. prot5408.

20. Kim J, Jang Y. [Investigation of tuberculosis in wildlife animals and dogs and cats residing on infected cattle and deer farms, and molecular characterization of Mycobacterium bovis isolated from animals]. Animal and Plant Quarantine Agency. (ed.).Annual Research Report. pp.p. 135–136. Animal and Plant Quarantine Agency;Gimcheon: 2014. Korean.
21. Kim N, Jang Y, Kim JK, Ryoo S, Kwon KH, Kang SS, Byeon HS, Lee HS, Lim YH, Kim JM. Complete genome sequence of Mycobacterium bovis clinical strain 1595, isolated from the laryngopharyngeal lymph node of South Korean cattle. Genome Announc. 2015; 3:e01124–15.

22. Kim N, Jang Y, Park SY, Song WS, Kim JT, Lee HS, Lim YH, Kim JM. Whole-genome squence of Mycobacterium bovis W-1171, isolated from the laryngopharyngeal lymph node of a wild boar in South Korea. Genome Announc. 2015; 3:e01464–15.

23. Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012; 62:716–721.

24. Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G, Wang Z, Rasko DA, McCombie WR, Jarvis ED, Adam M. Phillippy. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 2012; 30:693–700.
25. Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004; 5:R12.
26. Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007; 35:3100–3108.

27. Le Flèche P, Fabre M, Denoeud F, Koeck JL, Vergnaud G. High resolution, online identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2002; 2:37.
28. Lee SH, Lee WC. Epidemiological patterns and testing policies for bovine tuberculosis in the domestic cattle in Korea from 1961 to 2010. Japan J Vet Res. 2013; 61:19–23.
29. Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997; 25:955–964.

30. Ministry of Food and Drug Safety (KR). Livestock Product Sanitary Control. Act 12672. May 21, 2014.
31. Pirson C, Jones GJ, Steinbach S, Besra GS, Vordermeier HM. Differential effects of Mycobacterium bovis‒derived polar and apolar lipid fractions on bovine innate immune cells. Vet Res. 2012; 43:54.
32. Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009; 106:19126–19131.

33. Ruettger A, Nieter J, Skrypnyk A, Engelmann I, Ziegler A, Moser I, Monecke S, Ehricht R, Sachse K. Rapid spoligotyping of Mycobacterium tuberculosis complex bacteria by use of a microarray system with automatic data processing and assignment. J Clin Microbiol. 2012; 50:2492–2495.
34. Tatusova T, Ciufo S, Fedorov B, O'Neill K, Tolstoy I. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 2014; 42:D553–559.

35. Wiker HG, Harboe M. The antigen 85 complex: a major secretion product of Mycobacterium tuberculosis. Microbiol Rev. 1992; 56:648–661.
Fig. 1.
The entire genomic sequence of Mycobacterium bovis strain 1595. The scale is shown in megabases on the outer black circle. From the outside to the center: RNA features (ribosomal RNAs are shown in blue, and transfer RNAs are shown in red), genes on the forward strand, and genes on the reverse strand (colored according to the clusters of orthologous groups categories). The inner two circles show the GC ratio and GC skew. The GC ratio and GC skew shown in orange and red indicate positive values, respectively, and those shown in blue and green indicate negative values, respectively.
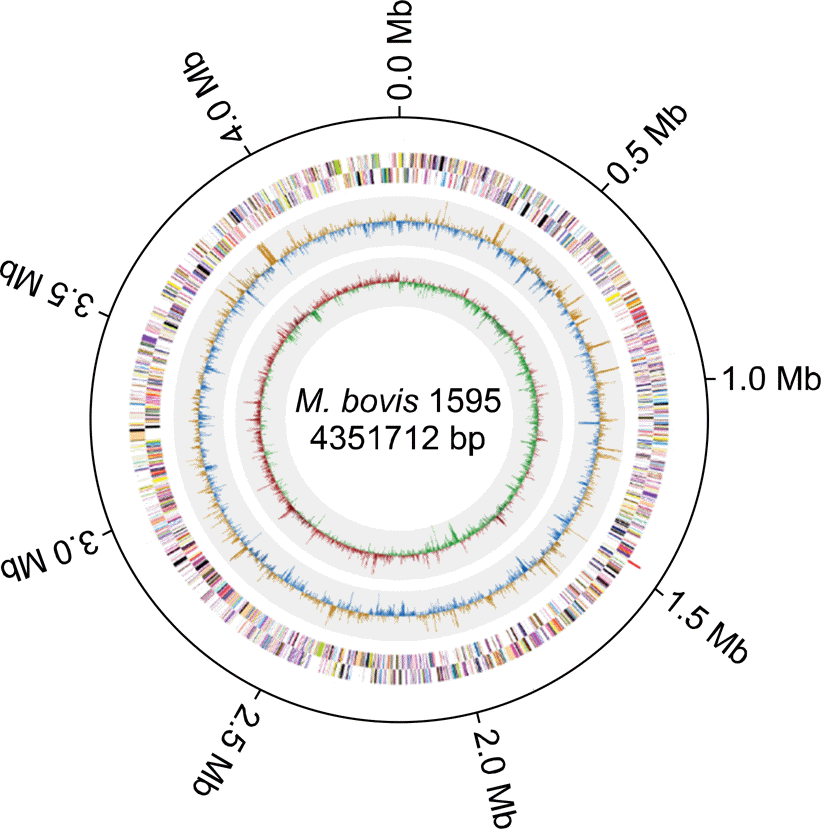
Fig. 2.
Genome tree based on the average nucleotide identity (ANI) values showing the relationships among Mycobacterium (M.) tuberculosis complex strains including M. bovis 1595. To convert the ANI value into a genetic distance, its complement to 1 was taken. From this pairwise distance matrix, an ANI tree was constructed using the unweighted pair group method and the arithmetic mean clustering method.
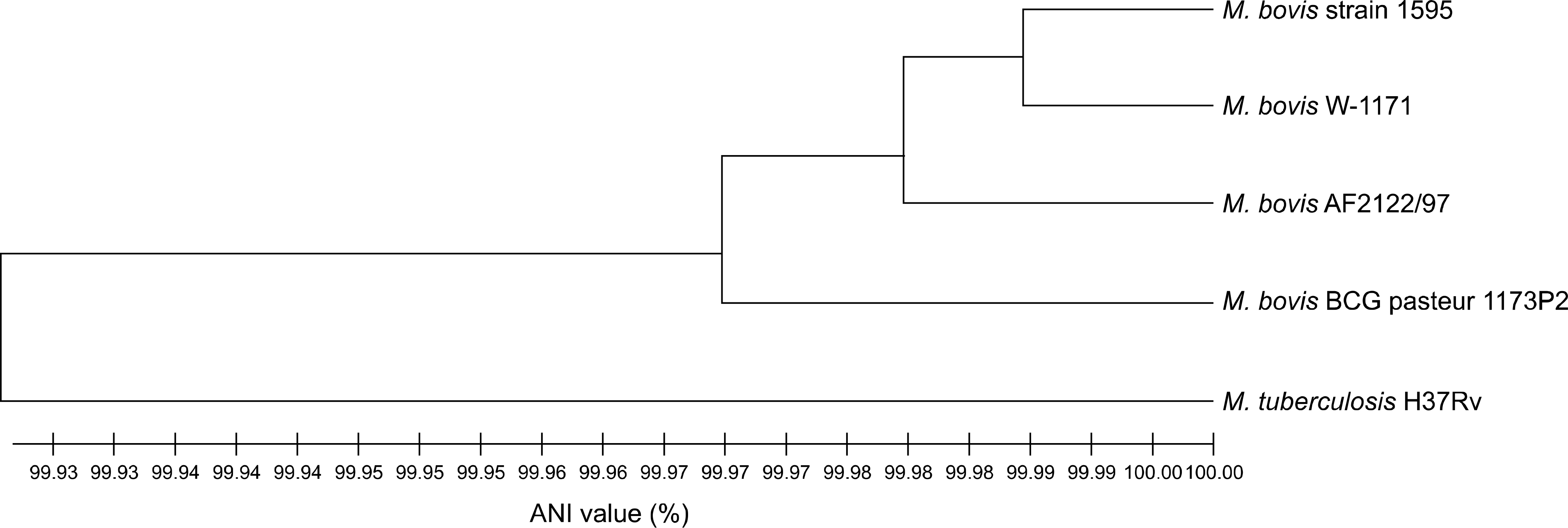
Fig. 3.
Nucleotide-based alignments with NUCmer. X-axis: Mycobacterium bovis strain 1595. Y-axis: (A) Mycobacterium bovis W-1171, (B) AF2122/97, (C) BCG Pasteur 1173P2, and (D) Mycobacterium tuberculosis H37Rv. Aligned segments are presented as dots or lines in the NUCmer alignment and were generated by the MUMmer plot script.
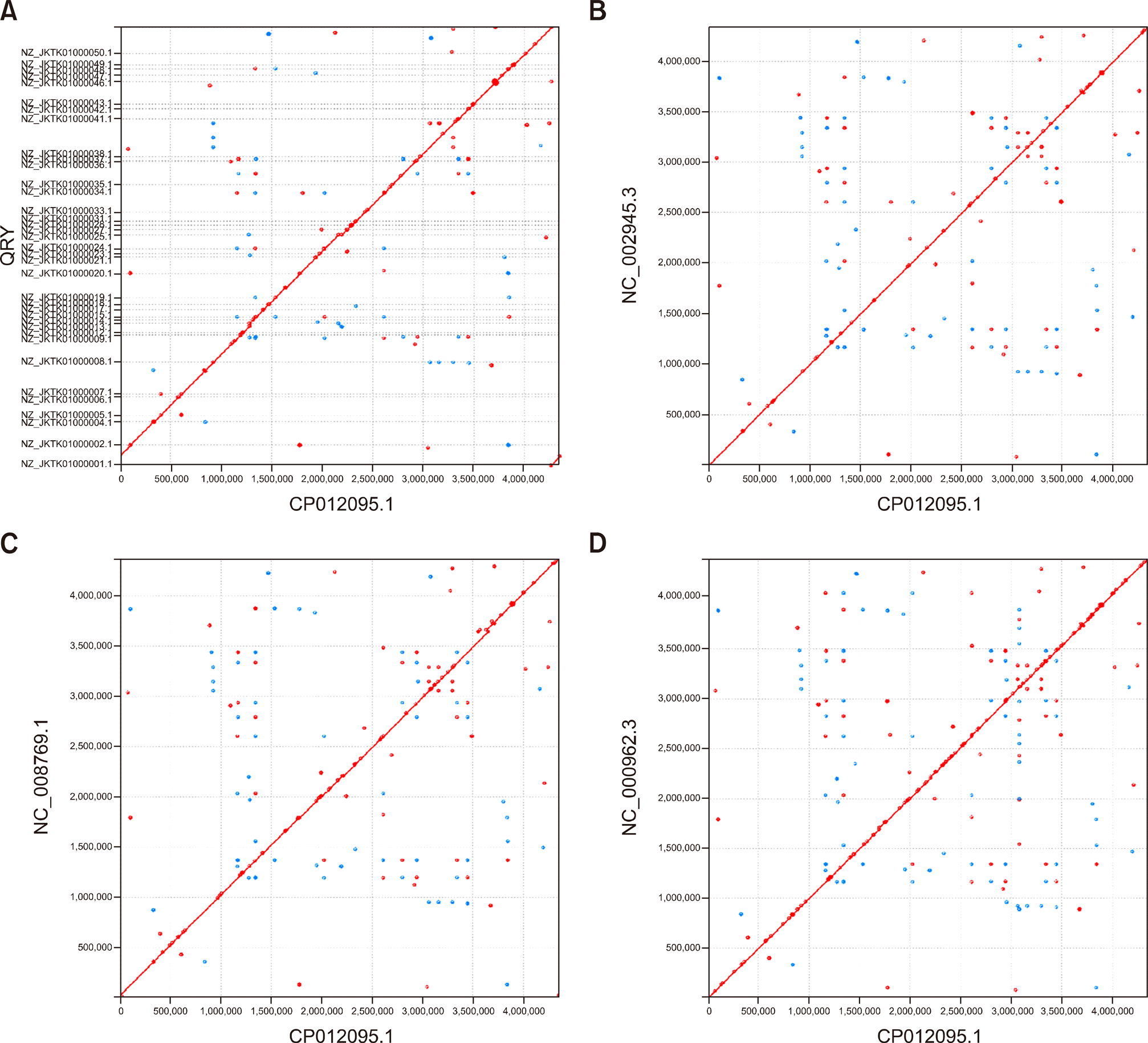
Table 1.
Clusters of orthologous groups (COGs) of the genome of Mycobacterium bovis strain 1595
Table 2.
Genomic characteristics of strains used in this study
Table 3.
Similarities of open reading frames (ORFs) of various Mycobacterium (M.) tuberculosis complex strains compared to M. bovis 1595 (n = 4,358)




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download