Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) has a high degree of genetic variation. In this study, we characterized the genetic variation and evolutionary relationships among circulating PRRSV strains in southern China. We analyzed 29 NSP2 strains and 150 ORF5 strains from clinical samples collected in southern China during 2007–2014. The alignment results showed that the nucleotide identity similarities of the two genes among these strains were 80.5%–99.7% and 80.9%–100%, respectively. Phylogenetic analysis based on the NSP2 gene showed that highly pathogenic (HP)-PRRSV was still the dominant virus in southern China from 2013 to 2014. Compared with reference strains CH-1a and VR-2332, the field strain 131101-GD-SHC, which shared high homology with JXA1-P170, had a novel 12 amino acid deletion at position 499–510. Phylogenetic analysis based on the ORF5 gene showed that HP-PRRSV, VR2332-like strains, and QYYZ-like strains were simultaneously circulating in southern China from 2007 to 2014, suggesting that, in recent years, the type 2 PRRSV was more diverse in southern China. In conclusion, mutations in the decoy epitope and primary neutralizing epitope could be markers of viral evolution and used to study evolutionary relationships among PRRSV strains in China.
Porcine reproductive and respiratory syndrome (PRRS), causes major economic losses to the swine industry [14]. PRRS was first discovered in 1987 [16] in the United States and identified in 1991 in the Netherlands [37]. The causative agent of PRRS is porcine reproductive and respiratory syndrome virus (PRRSV), which leads to reproductive disorders in late gestation sows and respiratory symptoms in pigs of all ages. In 2006, a large-scale outbreak of highly pathogenic PRRSV (HP-PRRSV) was reported for the first time in China and was characterized by high fever and mortality [332].
PRRSV is a small enveloped, nonsegmented, single-stranded, positive sense RNA virus in the order Nidovirales, family Arteriviridae [525]. The PRRSV genome is approximately 15 kb in length and contains at least ten open reading frames (ORFs) [25]. ORF1a and ORF1b are in the 5′-two-thirds of the viral genome and encode 13 functional nonstructural proteins. ORF2a, ORF2b, and ORFs 3–7 make up the 3′ terminal region and encode the viral structural proteins GP2, E, GP3, GP4, GP5, GP5a, M, and N [15]. NSP2, the largest replication protein in PRRSV, is highly variable and contains deletions, insertions, and point mutations [1241]. The HP-PRRSV strain that emerged in 2006 contains a discontinuous 30-amino acid deletion in the NSP2 gene [32]. ORF5 encodes the envelope glycoprotein GP5, which is the most variable structural gene in PRRSV and contains two to four glycosylation sites [11]. GP5 is crucial for infectivity [38] and viral entry into susceptible cells [33]. GP5 is a target for neutralization and elicits cellular and humoral immune responses. NSP2 and ORF5 are ideal candidates for tracking the evolution and molecular epidemiology of PRRSV because of their high degree of variability [2830].
PRRSV can be divided into two major genotypes that have approximately 60% nucleotide sequence identity: genotype 1 (European type) and genotype 2 (North American type) [537]. Representative strains of genotypes 1 and 2 are Lelystad virus and VR-2332, respectively. PRRSV can also be divided into conventional PRRSV (C-PRRSV), special mutant PRRSV (S-PRRSV) and HP-PRRSV strains [23]. C-PRRSV was first recognized in China in 1996 and the reference strains are CH-1a, HN1, and S1 [22]. HP-PRRSV was first recognized in 2006 and originated from a CH-1a-like C-PRRSV strain; JXA1, WUH4, and JXwn06 are representative strains in this group [12032]. S-PRRSV has specific mutation features and might be a virus strain generated by recombination. The reference strains of S-PRRSV are Em2007, SHB, HB-1(sh)/2002, and HB-2(sh)/ 2002, which are considered to be less virulent than JXA1, but more virulent than CH-1a [243].
Modified live-virus vaccines (MLV) are able to evoke both humoral and cell-mediated immune responses against homologous challenge. The available MLV vaccines against C-PRRSV are the CH-1R and VR2332 (Ingelvac PRRS MLV) vaccines, while the available vaccines for HP-PRRSV are JXA1-R, HuN4-F112, and TJM-92. The available MLV vaccines are effective in inducing protective immunity against homologous challenge [1318]. However, conventional vaccines have failed to protect against PRRS, especially after the outbreak of HP-PRRSV in 2006 [20]. The widespread use of the live attenuated vaccines such as the MLV RespPRRS, JXA1-R, HuN4-F112, and TJM might accelerate variation of PRRSV due to selective pressure [8]. Vaccine failure might be related to vaccine efficacy upon heterologous challenge [627]. In fact, a novel variant strain, GM2, has emerged and is likely a recombination between a MLV vaccine strain and the prototype Chinese field strain QYYZ [24]. The emergence of vaccine-like strains could make it more difficult to eradicate PRRSV.
Thus, it is important to describe the existing variation in, and evolutionary process of, PRRSV. In the present study, the evolution of PRRSV was analyzed, based on the ORF5 and NSP2 genes, by using strains collected on pig farms in southern China from 2007 to 2014. The genetic variation and phylogenetic relationships of the strains were compared with 45 Chinese reference strains and 15 foreign reference strains.
More than 1,000 clinical samples including serum, spleen, lung, kidney, sera, and lymph nodes were collected from symptomatic pigs exhibiting fever, dyspnea, reproductive failure, and acute death on farms in Guangdong province, China from 2007 to March 2015. Total RNA was extracted from tissue homogenates by using AxyPrep Body Fluid Viral DNA/RNA Miniprep Kit (Axygen, USA) according to the manufacturer's instructions. The remaining tissue samples were stored at −80℃.
To elucidate the genetic variation and molecular epidemiology of PRRSV, the NSP2 and ORF5 genes were amplified by using a one-step RT-PCR kit (Takara Bio, China) according to the manufacturer's instructions. The RT-PCR cycling sequence was as follows: 30 min cycle at 50℃, 3 min cycle at 94℃, followed by 34 cycles of denaturation at 94℃ for 30 sec, annealing at 55℃ for 30 sec, and extension at 72℃ for 1 min/kb. The samples were then incubated for an additional 10 min at 72℃. PCR products were examined by performing 1.0% agarose gel electrophoresis and were ligated into the pMD19-T vector (Takara Bio). Recombinant clones were sent to GENEWIZ (China) for sequencing.
The NSP2 and ORF5 sequences were edited with the DNASTAR Lasergene software (DNASTAR, USA) and independently used in sequence alignments and phylogenetic analysis. The ORF5 gene sequences from 44 reference PRRSV sequences were obtained from GenBank, aligned with 150 strains by using the Clustal W program [7], and the percentage nucleotide sequence similarities were assessed further. Twenty-nine NSP2 sequences were aligned with 33 reference strains. Phylogenetic trees were constructed from aligned nucleotide sequences based on the NSP2 and ORF5 genes by using the maximum likelihood method in MEGA 5 software [31].
More than 1,000 tissue samples were collected throughout Guangdong province, China including areas in and near the cities of Yunfu, Qingyuan, Foshan, Jiangmen, Yanjiang, Heshan, and Shaoguan. Of 675 sequences identified, 150 ORF5 sequences and 29 NSP2 sequences were selected for sequence alignments and phylogenetic analysis after removing all duplicate sequences. Temporal and geographical distributions were taken into consideration during the sequence selection process. The strain names, isolation dates, and geographical regions are presented in more detail in Supplementary Table 1.
To better understand the molecular evolution of PRRSV we characterized the phylogenetic and amino acid similarities among the sequenced strains and the reference PRRSV strains (Supplementary Table 2). Pairwise comparisons showed that the 29 NSP2 sequences shared 80.5%–99.7% and 73.5%–100.0% similarities at the nucleotide and amino acid levels, respectively. The amino acid similarities, when compared to the HP-PRRSV strain JXA1 and the genotype 2 strain VR-2332, ranged from 76.8%–98.3% and 74.0%–99.4%, respectively. Twenty-five NSP2 sequences were 950 amino acids in length, which is similar to JXA1; however, two NSP2 sequences were 980 amino acids in length, which was similar to VR-2332, and the remaining two NSP2 sequences were 938 and 986 amino acids in length.
To better understand the genetic relationships among PRRSV strains, phylogenetic analysis was conducted using the 29 NSP2 sequences and the representative sequences. The phylogenetic tree indicated that all 29 NSP2 sequences belonged to genotype 2 of PRRSV and could be classified into four of the five subgroups (Fig. 1). Sequence 131005-GD-CWC3 was located in subgroup 5 along with the representative strains VR-2332, BJ-4, and MLV RespPRRS, while 1205-GD-TTC and 131010-GD-HTB were clustered in subgroup 2 and subgroup 4, respectively. The remaining 26 isolates were clustered into multiple branches of subgroup 1 along with the representative strains JXA1, HUB1, and JXA1 P170.
To identify regions where deletions had occurred, the 29 isolated NSP2 genes were compared with 10 reference isolates from China and other countries. The nucleotide and deduced amino acid sequence analysis revealed that, compared to genotype 2 prototype VR-2332 and Chinese early strain CH-1a, 27 of the 29 NSP2 sequences contained the same 30-amino acid deletion at position 481 and position 533–561 as observed in JXA1 and other HP-PRRSV strains (panel A in Fig. 2). Alignment of the predicted amino acid sequences of the NSP2 gene indicated that PRRSVs contained various deletions and insertions that led to NSP2 proteins ranging from 938–986 amino acid residues. However, compared with CH-1a, 131101-GD-SHC showed an additional novel 12 amino acid deletion at position 499–510. Notably, when compared with CH-1a and VR-2332, the 131010-GD-HTB sequence contained the same 36 amino acid insertions at position 812–813 (panel B in Fig. 2) as that in QYYZ which increased the protein length from 950 to 986 amino acids.
We next compared the amino acid and nucleotide similarities among isolates at the ORF5 gene. The 150 strains shared a nucleotide identity ranging from 80.9%–100% and an amino acid homology ranging from 76.6%–100%. In addition, the 150 strains had 61.3%–65.0% nucleotide identity and 82.3%–100% amino acid homology with the genotype 1 Lelystad virus reference strain and 83.3%–95.7% nucleotide identity and 81.9%–99.7% amino acid homology with the genotype 2 VR2332 reference strain. When compared to Chinese PRRSV strains, the isolates had 83.3%–95.7% nucleotide identity and 81.9%–99.7% amino acid homology with CH-1a, and 81.9%–99.8% nucleotide identity and 79.6%–99.5% amino acid homology with JXA1 (HP-PRRSV).
To elucidate the genetic relationships and evolution of PRRSV in China, 150 complete ORF5 sequences were aligned with 49 reference strains, and a phylogenetic tree based on ORF5 genes was constructed (Fig. 3). All of the Chinese PRRSV strains were grouped within the genotype 2 PRRSVs, which can be divided into five subgroups. The nucleotide and amino acid identity among each subgroup of the Chinese PRRSV strains based on ORF5 genes are displayed in Table 1. Of these isolates, 82 Chinese PRRSVs clustered in subgroup 1 were most closely related to JXA1 and had 95.7%–99.8% nucleotide similarity. The 7 ORF5s that belonged to subgroup 2 were closely related to Em2007. The PRRSV strains in subgroup 5 appeared between 2010 and 2015 and are closely related to VR2332 and the MLV RespPRRS with 96.5%–100% and 96.5%–99.8% nucleotide similarity, respectively. Subgroup 4 contained 32 PRRSV strains from 2011 to early 2015 that were closely related to QYYZ and GM2. However, no strain was clustered into subgroup 3, which was considered to be a C-PRRSV.
GP5 is one of the most variable structural proteins in the PRRSV genome. We investigated the degree of genetic divergence among 150 strains and 44 reference sequences, based on the reported functional domains of GP5. Based on an amino acid analysis of GP5, four hypervariable regions were observed at positions 3–16, 24–39, 92–104, and 185–200.
We first examined the primary neutralizing epitope (PNE) S37H(F/L)QLIYNL and compared it to CH-1a (Table 2). All of the PRRSV sequences in subgroup 1 and subgroup 2 had the same amino acid mutation (F39→I39), and all of the strains in subgroup 4 had two amino acid mutations (H38→Y38, F39→S39). We next compared the decoy epitope (A/V)27LVN to CH-1a. Twenty-one strains in subgroup 1 had one mutation (V29→A29) in the decoy epitope. All of the strains in subgroup 5 had the same mutation (V29→A29), which is also shared with 130123-QYYZC, 120301-GD-QYYZC, 150112-GD-NMB, and 150305-GD-HYB in subgroup 4. Furthermore, all strains in subgroup 4, including QYYZ and GM2, had one amino acid mutation (N30→S30).
We also analyzed potential N-glycosylation sites (NGSs; NXS/T) in the GP5 extra-virion domain. The number and distribution of the potential NGSs in the GP5 domain are shown in Table 3. Glycosylation of the envelope protein is likely to be related to immune evasion. Of the 82 HP-PRRSV strains, 58 of the ORF5s in subgroup 1 had four potential NGSs that were similar to those in JXA1. Most of strains in subgroup 2 had four potential NGSs, only 130302-GD-DBEC had three. Twenty-six of the 29 strains in subgroup 5 had four potential NGSs (N30, N33, N44, and N51), and two of the remaining strains had three potential NGSs, while 150125-GD-XWC2 had an additional NGS (N81). Most of the strains in subgroup 4 had three potential NGSs except for two strains, 140107-GD-SHC and 110901-GD-SWKF, which had only two NGSs. Furthermore, the potential NGS at position 51 occurred in all strains.
The PRRSV is a pathogen that has resulted in enormous losses in the global swine industry since its first occurrence in the 1980s. In this study, we describe the genetic variation and evolutionary relationships between PRRSV strains circulating in southern China between 2007 and 2014 based on the sequences of NSP2 and ORF5. NSP2 was chosen because it contains the highest level of genetic diversity among the nonstructural proteins. Based on the NSP2 sequences, all 29 strains belonged to genotype 2, which can be further subdivided into five subgroups. Of the 29 strains, 26 belonged to subgroup 1, which is represented by the reference strain JXA1, and one strain each were clustered with subgroups 2, 4, and 5 represented by reference strains HB-1(sh), QYYZ, and VR-2332, respectively. The majority of Chinese PRRSV strains were grouped with genotype 2 PRRSVs, which have been systematically classified into lineages and sublineages [30]. Subgroup 1 was comprised of HP-PRRSV strains that have emerged since 2006. JXA1, WUH4, and JXwn06, which are representative strains in this subgroup, were clustered into sublineage 8.7. Strains in subgroup 3, which are considered to be conventional PRRSVs, were also clustered into sublineage 8.7. The intermediate isolates in subgroup 2, such as SHB, Em2007, HB-1(sh)/2002, and HB-2(sh)/ 2002, are considered to be less virulent than JXA1, but more virulent than CH-1a [243]. According to previous study [30], strains in subgroup 4 and 5 belonged to lineage 3 and sublineage 5.1, respectively.
Interestingly, 27 of the 29 strains contained the same 30-amino acid deletion in NSP2 at position 481 and position 533–561 as has been reported in JXA1 and other HP-PRRSV strains that have emerged since 2006 [32]. Because the virulence of the PRRSV was thought to be related to several factors, whether the 27 PRRSV strains with the 30 amino acid deletion are highly pathogenic requires further investigation.
In recent years, some novel PRRSV strains with amino acid deletions or insertions have been reported. Recently, one strain with a novel continuous 49 amino acid deletion at position 473–521 [17], some strains with deletions at position 503, 593–595 [34], and some strains with deletion at position 495–513 [35] have been reported. In this study, in contrast to the CH-1a reference strain, a 12 amino acid deletion in the NSP2-coding region was observed at position 499–510 of 131101-GD-SHC. That novel deletion was first observed in a PRRSV strain in Guangdong province. A 36 amino acid insertion following the amino acid at position 816 was discovered in 131010-GD-HTB, similar to insertions in QYYZ and GM2. All of the insertions and deletions observed in the 29 field isolates were located at the hypervariable region between amino acids 150–850 in NSP2. However, the virulence and pathogenicity of the HP-PRRSVs that have emerged in China are unrelated to the 30-amino-acid discontinuous deletion in NSP2 [44]. Strain 131010-GD-HTB, which was clustered into subgroup 4, also had the same 30 amino acid discontinuous deletion in NSP2, suggesting that a strain with such a deletion might be not necessarily be a HP-PRRSV. Similarly, several studies have reported that this deletion can no longer be defined as a molecular marker of HP-PRRSV because of the large number of low-pathogenicity strains that contain the same 30 amino acid deletion in NSP2 [101822].
The ORF5 gene is also a highly variable gene in PRRSVs and has an important role in humoral immunity. As with NSP2, all 150 ORF5 sequences belonged to genotype 2. All of the different yearly sequenced strains clustered into subgroups irregularly and did not exhibit any tendencies. The results of this study suggest that there is no clear relationship, geographically or temporally, among the Chinese PRRSV strains from 2007 to 2014. The 150 ORF5 strains shared 80.9%–100% identity at the nucleotide level and can be further divided into five subgroups, suggesting diversity among the Chinese PRRSVs from 2007 to 2014. However, only seven strains fell into subgroup 2. Of the 150 sequences, 82 showed high homology to the HP-PRRSV reference strains, which indicates that HP-PRRSV were still dominant in southern China from 2007 to 2014. Moreover, the phylogenetic tree showed that the HP-PRRSVs were genetically quite distant from the MLV vaccine RespPRRS. Most of the affected pigs had been vaccinated with MLV vaccine, which might indicate that the current PRRS vaccines do not provide sufficient protection against HP-PRRSV.
It is noteworthy that 32 strains were clustered into subgroup 4 and shared high homology with GM2, a likely vaccine recombinant strain [24]. The re-emerged viruses in subgroup 4 were closely related to the 2004 isolates from Hong Kong and might also be considered to be classical PRRSVs. Genetic recombination has previously occurred between attenuated vaccine strains of genotype 2 PRRSV in vitro [42], suggesting that recombination is probably a genetic mechanism with an important role in PRRSV evolution. Thus, we hypothesize that the strains in subgroup 4 might also be recombinants between field strains and vaccine strains. However, a more thorough analysis of natural genetic recombination should be performed by undertaking a detailed bioinformatics study. Seven strains that shared high nucleotide similarities with reference strains SHB and HB-1(sh) were considered S-PRRSVs. This could indicate that Chinese PRRSV strains may have evolved through gradual genomic variation. Subgroup 5 comprised 29 PRRSV strains that were highly homologous to each other and closely related to the MLV RespPRRS vaccine and its parent, the VR-2332 strain. The occurrence of these strains is presumably associated with the extensive use of the attenuated modified live PRRS vaccine.
We hypothesize that HP-PRRSV, S-PRRSV, and QYYZ-like strains have coexisted and spread widely in southern China in recent years, which is counter to suppositions in previous work. Some authors have proposed that extensive substitutions have occurred in the signal peptide, PNE, and decoy epitope of ORF5 [2040]. However, even if ORF5 is one of the most variable genes in the PRRSV genome, deletions and insertions in this region have rarely been recognized in recent studies. The amino acid analysis in this study indicates that 25 strains from subgroup 1 and all of the strains in subgroups 4 and 5 contain a one or two amino acid mutation in the decoy epitope. The PNE, S37H(F/L)QLIYNL45, is a critical epitope for viral neutralization, and amino acids H38L39/F39 are considered as critical sites for neutralizing activity [4]. None of the strains in subgroup 5 contained a mutation in the PNE. Surprisingly, all of the strains in subgroups 1, 2, and 4 contained a substitution from leucine or phenylalanine to isoleucine or serine; moreover, the strains in subgroup 4 even had an additional amino acid mutation (H38→Y38). These substitutions may affect the conformation required for Ab binding [29]. GP5 of PRRSV possesses two to four potential NGSs [9]. The NGSs in GP5 are reported to be involved in evasion from neutralizing antibodies and in minimizing the immunogenicity of the nearby neutralization epitope [4]. Analysis of the potential NGSs indicated that most of the strains in subgroups 1 and 5 contained four NGSs, but 30 out of 32 strains in subgroup 4 had three NGSs. Notably, strains 140107-GD-SHC and 110901-GD-SWKF in subgroup 4 had only two NGSs. All of the strains shared conserved potential NGSs at residue 51, which have been well characterized [21,263639]. Nevertheless, potential NGSs at residue 44 were lost in three strains of subgroup 1, which might increase the sensitivity of a mutant virus to convalescent-phase serum samples [36]. Two unique amino acid mutations were found at position 30 (N→S) and position 33 (N→G) in most strains in subgroup 4. These substitutions might contribute to the loss of two potential NGSs in GP5. In this study, the potential N-glycosylation results for subgroup 5 were markedly different from those in a previous study, which showed that the number of potential NGSs of Chinese isolates had significantly increased with time [19]. Additional research is required to clarify the interaction between the NGSs and neutralization epitope.
In conclusion, the PRRSV strains collected in southern China from 2007 to 2014 were highly similar to HP-PRRSVs and VR2332. The amino acid analysis of the 150 ORF5 and 29 NSP2 strains from 2007 to 2014 has revealed that the Chinese PRRSV has undergone gradual variation and evolution accumulation processes in recent years. Thirty-two strains were clustered into subgroup 4, indicating that QYYZ-like strains might widely re-disseminate in the field. PRRSV eradication will be made more difficult by the presence of simultaneously circulating genetic variants and recombination between strains. Thus, it is particularly important to monitor the prevalence of PRRS and to elucidate the variations among circulating PRRSV strains in China. Such investigations can provide reference data for selecting PRRS vaccine strains. This study provided new information about the molecular variation and evolutionary relationships between PRRSV strains currently disseminated in southern China, but further work is needed.
Figures and Tables
Fig. 1
Phylogenetic tree comparing PRRSV field strains and reference strains based on NSP2 gene sequences. Field PRRSV strains were collected between 2012 and 2014. The NSP2 gene (n = 29) was sequenced and compared to published reference strains in GenBank (n = 35; filled circles). The reliability of the tree was assessed by performing a bootstrap analysis with 1000 replications. The distinct subgroups are marked with different colors as indicated on the tree.

Fig. 2
The deduced amino acid sequence alignment of the NSP2 gene from PRRSV field strains and reference strains. (A) The deduced amino acid sequence alignment of the NSP2 at position 470–564 from PRRSV field strains and reference strains. (B) The deduced amino acid sequence alignment of the NSP2 at position 810–900 from PRRSV field strains and reference strains. The amino acid sequence of the complete NSP2 gene sequence from field strains (n = 29) collected between 2012 and 2014 was compared to published reference strains (n = 10). Solid dots indicate amino acids identical to the CH-1a sequence. Dashes indicate gaps. The red boxed residues indicate deletion regions. The blue boxed residues indicate insertion regions.

Fig. 3
Phylogenetic tree comparing PRRSV field strains and reference strains based on the ORF5 gene. Field PRRSV strains were collected between 2011 and 2014. The ORF5 gene (n = 150) was sequenced and compared to published reference strains in GenBank (n = 49; filled circles). The reliability of the tree was assessed by performing a bootstrap analysis with 1000 replications. The distinct subgroups are marked with different colors as indicated on the tree.
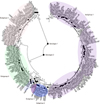
Table 1
Genotyping based on ORF5 in porcine reproductive and respiratory syndrome virus (PRRSV) strains collected between 2007 and 2015

Acknowledgments
The study was financially supported by a grant of Science and Technology Specific Projects of Guangdong Province (2014A020208093) from the Guangdong Science and Technology Department of China.
References
1. An TQ, Tian ZJ, Xiao Y, Li R, Peng JM, Wei TC, Zhang Y, Zhou YJ, Tong GZ. Origin of highly pathogenic porcine reproductive and respiratory syndrome virus, China. Emerg Infect Dis. 2010; 16:365–367.

2. An TQ, Tian ZJ, Zhou YJ, Xiao Y, Peng JM, Chen J, Jiang YF, Hao XF, Tong GZ. Comparative genomic analysis of five pairs of virulent parental/attenuated vaccine strains of PRRSV. Vet Microbiol. 2011; 149:104–112.

3. An TQ, Zhou YJ, Liu GQ, Tian ZJ, Li J, Qiu HJ, Tong GZ. Genetic diversity and phylogenetic analysis of glycoprotein 5 of PRRSV isolates in mainland China from 1996 to 2006: coexistence of two NA-subgenotypes with great diversity. Vet Microbiol. 2007; 123:43–52.

4. Ansari IH, Kwon B, Osorio FA, Pattnaik AK. Influence of N-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J Virol. 2006; 80:3994–4004.

5. Benfield DA, Nelson E, Collins JE, Harris L, Goyal SM, Robison D, Christianson WT, Morrison RB, Gorcyca D, Chladek D. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J Vet Diagn Invest. 1992; 4:127–133.

6. Cano JP, Dee SA, Murtaugh MP, Pijoan C. Impact of a modified-live porcine reproductive and respiratory syndrome virus vaccine intervention on a population of pigs infected with a heterologous isolate. Vaccine. 2007; 25:4382–4391.

7. Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003; 31:3497–3500.

8. Costers S, Lefebvre DJ, Van Doorsselaere J, Vanhee M, Delputte PL, Nauwynck HJ. GP4 of porcine reproductive and respiratory syndrome virus contains a neutralizing epitope that is susceptible to immunoselection in vitro. Arch Virol. 2010; 155:371–378.

9. Dea S, Gagnon CA, Mardassi H, Pirzadeh B, Rogan D. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch Virol. 2000; 145:659–688.

10. Du Y, Lu Y, Qi J, Wu J, Wang G, Wang J. Complete genome sequence of a moderately pathogenic porcine reproductive and respiratory syndrome virus variant strain. J Virol. 2012; 86:13883–13884.

11. Faaberg KS, Even C, Palmer GA, Plagemann PG. Disulfide bonds between two envelope proteins of lactate dehydrogenase-elevating virus are essential for viral infectivity. J Virol. 1995; 69:613–617.

12. Fang Y, Kim DY, Ropp S, Steen P, Christopher-Hennings J, Nelson EA, Rowland RRR. Heterogeneity in Nsp2 of European-like porcine reproductive and respiratory syndrome viruses isolated in the United States. Virus Res. 2004; 100:229–235.

13. Han W, Wu JJ, Deng XY, Cao Z, Yu XL, Wang CB, Zhao TZ, Chen NH, Hu HH, Bin W, Hou LL, Wang LL, Tian KG, Zhang ZQ. Molecular mutations associated with the in vitro passage of virulent porcine reproductive and respiratory syndrome virus. Virus Genes. 2009; 38:276–284.

14. Holtkamp DJ, Kliebenstein JB, Neumann EJ, Zimmerman JJ, Rotto HF, Yoder TK, Wang C, Yeske PE, Mowrer CL, Haley CA. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J Swine Health Prod. 2013; 21:72–84.
15. Huang C, Zhang Q, Feng WH. Regulation and evasion of antiviral immune responses by porcine reproductive and respiratory syndrome virus. Virus Res. 2015; 202:101–111.

16. Keffaber KK. Reproductive failure of unknown etiology. Am Assoc Swine Pract Newsl. 1989; 1:1–9.
17. Leng CL, Tian ZJ, Zhang WC, Zhang HL, Zhai HY, An TQ, Peng JM, Ye C, Sun L, Wang Q, Sun Y, Li L, Zhao HY, Chang D, Cai XH, Zhang GH, Tong GZ. Characterization of two newly emerged isolates of porcine reproductive and respiratory syndrome virus from Northeast China in 2013. Vet Microbiol. 2014; 171:41–52.

18. Leng X, Li Z, Xia M, He Y, Wu H. Evaluation of the efficacy of an attenuated live vaccine against highly pathogenic porcine reproductive and respiratory syndrome virus in young pigs. Clin Vaccine Immunol. 2012; 19:1199–1206.

19. Li B, Fang L, Guo X, Gao J, Song T, Bi J, He K, Chen H, Xiao S. Epidemiology and evolutionary characteristics of the porcine reproductive and respiratory syndrome virus in China between 2006 and 2010. J Clin Microbiol. 2011; 49:3175–3183.

20. Li Y, Wang X, Bo K, Wang X, Tang B, Yang B, Jiang W, Jiang P. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the Mid-Eastern region of China. Vet J. 2007; 174:577–584.

21. Li Y, Wang X, Jiang P, Wang X, Chen W, Wang X, Wang K. Genetic variation analysis of porcine reproductive and respiratory syndrome virus isolated in China from 2002 to 2007 based on ORF5. Vet Microbiol. 2009; 138:150–155.

22. Li Y, Xue C, Wang L, Chen X, Chen F, Cao Y. Genomic analysis of two Chinese strains of porcine reproductive and respiratory syndrome viruses with different virulence. Virus Genes. 2010; 40:374–381.

23. Liu C, Ning Y, Xu B, Gong W, Zhang D. Analysis of genetic variation of porcine reproductive and respiratory syndrome virus (PRRSV) isolates in central China. J Vet Med Sci. 2016; 78:641–648.

24. Wenhui L, Zhongyan W, Guanqun Z, Zhili L, JingYun M, Qingmei X, Baoli S, Yingzuo B. Complete genome sequence of a novel variant porcine reproductive and respiratory syndrome virus (PRRSV) strain: evidence for recombination between vaccine and wild-type PRRSV strains. J Virol. 2012; 86:9543.

25. Meulenberg JJ, de Meijer EJ, Moormann RJ. Subgenomic RNAs of Lelystad virus contain a conserved leader-body junction sequence. J Gen Virol. 1993; 74:1697–1701.

26. Meulenberg JJ, Hulst MM, de Meijer EJ, Moonen PL, den Besten A, de Kluyver EP, Wensvoort G, Moormann RJ. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 1993; 192:62–72.

27. Murtaugh MP, Xiao Z, Zuckermann F. Immunological responses of swine to porcine reproductive and respiratory syndrome virus infection. Viral Immunol. 2002; 15:533–547.

28. Nilubol D, Tripipat T, Hoonsuwan T, Tipsombatboon P, Piriyapongsa J. Genetic diversity of the ORF5 gene of porcine reproductive and respiratory syndrome virus (PRRSV) genotypes I and II in Thailand. Arch Virol. 2013; 158:943–953.

29. Plagemann PG. The primary GP5 neutralization epitope of North American isolates of porcine reproductive and respiratory syndrome virus. Vet Immunol Immunopathol. 2004; 102:263–275.

30. Shi M, Lam TTY, Hon CC, Murtaugh MP, Davies PR, Hui RKH, Li J, Wong LTW, Yip CW, Jiang JW, Leung FCC. Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses. J Virol. 2010; 84:8700–8711.

31. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28:2731–2739.

32. Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One. 2007; 2:e526.

33. Van Breedam W, Van Gorp H, Zhang JQ, Crocker PR, Delputte PL, Nauwynck HJ. The M/GP5 glycoprotein complex of porcine reproductive and respiratory syndrome virus binds the sialoadhesin receptor in a sialic acid-dependent manner. PLoS Pathog. 2010; 6:e1000730.

34. Wang FX, Qin LT, Liu Y, Liu X, Sun N, Yang Y, Chen T, Zhu HW, Ren JQ, Sun YJ, Cheng SP, Wen YJ. Novel Nsp2 deletion based on molecular epidemiology and evolution of porcine reproductive and respiratory syndrome virus in Shandong Province from 2013 to 2014. Infect Genet Evol. 2015; 33:219–226.

35. Wang X, He K, Zhang W, Zhou Z, Mao A, Yu Z. Genetic diversity and phylogenetic analysis of porcine reproductive and respiratory syndrome virus isolates in East China. Infect Genet Evol. 2014; 24:193–201.

36. Wei Z, Lin T, Sun L, Li Y, Wang X, Gao F, Liu R, Chen C, Tong G, Yuan S. N-linked glycosylation of GP5 of porcine reproductive and respiratory syndrome virus is critically important for virus replication in vivo. J Virol. 2012; 86:9941–9951.

37. Wensvoort G, Terpstra C, Pol JMA, ter Laak EA, Bloemraad M, de Kluyver EP, Kragten C, van Buiten L, den Besten A, Wagenaar F, Broekhuijsen JM, Moonen PLJM, Zetstra T, de Boer EA, Tibben HJ, de Jong MF, van 't Veld P, Greenland GJR, van Gennep JA, Voets MTh, Verheijden JHM, Braamskamp J. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q. 1991; 13:121–130.

38. Wissink EH, van Wijk HA, Kroese MV, Weiland E, Meulenberg JJ, Rottier PJ, van Rijn PA. The major envelope protein, GP5, of a European porcine reproductive and respiratory syndrome virus contains a neutralization epitope in its N-terminal ectodomain. J Gen Virol. 2003; 84:1535–1543.

39. Xie J, Zhu W, Chen Y, Wei C, Zhou P, Zhang M, Huang Z, Sun L, Su S, Zhang G. Molecular epidemiology of PRRSV in South China from 2007 to 2011 based on the genetic analysis of ORF5. Microb Pathog. 2013; 63:30–36.

40. Yin G, Gao L, Shu X, Yang G, Guo S, Li W. Genetic diversity of the ORF5 gene of porcine reproductive and respiratory syndrome virus isolates in Southwest China from 2007 to 2009. PLoS One. 2012; 7:e33756.

41. Yuan S, Mickelson D, Murtaugh MP, Faaberg KS. Complete genome comparison of porcine reproductive and respiratory syndrome virus parental and attenuated strains. Virus Res. 2001; 79:189–200.

42. Yuan S, Nelsen CJ, Murtaugh MP, Schmitt BJ, Faaberg KS. Recombination between North American strains of porcine reproductive and respiratory syndrome virus. Virus Res. 1999; 61:87–98.

Supplementary Materials
Supplementary data is available at http://www.vetsci.org only.




 Citation
Citation Print
Print




 XML Download
XML Download