Abstract
There are high levels of co-incidence of porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus type 2 (PCV2) in porcine tissue. This study established a duplex nested reverse transcriptase polymerase chain reaction (RT-PCR) method that targets the genomic RNA of type 2 PRRSV and the mRNA of PCV2 in infected tissues. The method amplified discriminative bands of 347 bp and 265 bp specific for type 2 PRRSV and PCV2, respectively. The limits of detection of the duplex nested RT-PCR were 101.5 TCID50/mL for type 2 PRRSV and 102 infected cells/mL for PCV2. The kappa statistic, which measures agreement between methods, was 0.867, indicating a good level of agreement. This RNA-based duplex RT-PCR approach can be another way to detect type 2 PRRSV and PCV2 simultaneously and with improved convenience.
Porcine reproductive and respiratory syndrome virus (PRRSV) is one of the main pathogens causing respiratory and reproductive diseases in swine. PRRSV is a small, enveloped, positive single-stranded RNA virus belonging to the family Arteriviridae of order Nidovirales. In pigs, PRRSV is detected in cells of macrophage lineage located within tissues of lung, heart, lymph nodes, tonsil, nasal turbinate, thymus, spleen, intestine, kidney, liver, adrenal gland, brain, and testes [512]. Porcine circovirus 2 (PCV2) is a small, non-enveloped, circular, single-stranded DNA virus of the family Circoviridae [10]. PCV2 has been associated with a clinical condition known as postweaning multisystemic wasting syndrome (PMWS), which is observed in pigs shortly after weaning [2]. PCV2 has been detected in several tissues and organs such as lung, tonsil, lymph node, thymus, spleen, intestine, kidney, liver, serum, salivary gland, and testes after experimental infection [37].
Recently, dual infection of PRRSV and PCV2 was regarded as one of the most common potential causes of PMWS [11], and experimental production of PMWS was observed when the pigs were dually infected with PRRSV and PCV2 [16]. In Korea, among 105 pigs with porcine respiratory disease complex, 85 and 66 pigs were shown to be infected with PCV2 and PRRSV, respectively [8]. Based on these previous reports, PRRSV and PCV2 dual infection was increased and related to the wasting diseases, and detection of these two viruses by diagnostic laboratories was deemed essential. There are high co-incidences of PRRSV and PCV2 in tissues, and pooled tissue samples of pigs with doubtful viral status could be used to detect dual infections of PRRSV and PCV2. However, there is some inconvenience associated with extracting both RNA and DNA from a pooled tissue sample due to genomic differences, although genomic DNA and mRNA of PCV2 have similar patterns of tissue distribution [15]. Therefore, by targeting the genomic RNA of PRRSV and the mRNA of capsid protein in PCV2-infected tissues, a duplex nested reverse transcriptase polymerase chain reaction (RT-PCR) method could be used to detect both PRRSV and PCV2 from the total RNA extracted from pooled tissue samples.
In this study, the primers for detection of type 2 PRRSV followed those reported by Kono et al. [9], of which the 668 bp and 347 bp products were amplified by outer (N21/N26) and inner (N22/N24) primers, respectively. For detection of PCV2 via mRNA, the outer primers were CapF/CapR which amplified a 594 bp PCR fragment [14], and the newly designed inner primers were Capn1: GGTAAAAGCAAATGGGCTGC and Capn2: CAGTTGAGGAGTACCATTCCAAC which amplified a 265 bp PCR product. Total RNA was extracted by using TRIzol LS (Invitrogen, USA) according to the manufacturer's instructions. The RNA was converted into cDNA by using random hexamers (50 pmol/µL) and commercial M-MLV reverse transcriptase kit (Invitrogen) following the manufacturer's protocol.
A commercial i-StarMaster mix PCR Kit (iNtRON Biotechnology, Korea) was used for PCR reactions. A 0.5 µL aliquot of each primer (N21/N26 for PRRSV and CapF/CapR for PCV2, 10 pmol/µL each) and 16 µL of i-StarMaster mix solution were mixed with 2 µL of the reverse transcripted products, resulting in total volume of 20 µL. The outer PCR was performed with denaturing at 94℃ for 5 min and cycling 35 times at 94℃ for 30 sec, 58℃ for 30 sec and 72℃ for 40 sec, and final elongating at 72℃ for 7 min. The nested PCR was performed with same protocol as the above mentioned except for the use of nested primer sets (N22/N24 for type 2 PRRSV and Capn1/Capn2 for PCV2) and 30 cycles with an annealing temperature of 55℃. The resultant nested PCR product was electrophoresed in 1.5% agarose gel and photographed. To investigate cross-reactivity, several controls were used, such as: (1) type 2 PRRSV isolate, (2) tissue sample co-infected with type 2 PRRSV and PCV2, (3) PCV2-infected PK15 cells, and (4) PCV1-infected PK15 cells. The detection limits of this duplex nested RT-PCR were determined by using 10-fold dilutions of a mixture of 105 PCV2-infected PK15 cells/mL (field isolate A4275) and 105.5 TCID50/mL type 2 PRRSV (field isolate CP07-401-9).
As shown in Fig. 1, the duplex nested RT-PCR successfully amplified target bands of 265 bp for PCV2 (lane 5) and 347 bp for type 2 PRRSV (lane 6). Non-specific amplification was not visualized when the templates were prepared from either PCV1-infected PK15 cells (lanes 1–3), PCV1-free PK15 cells (lane 4), or MARC-145 cells (lane 8). Two specific PCR products were observed from a tissue that was co-infected by type 2 PRRSV and PCV2 (lane 7). The limits of detection of the duplex nested RT-PCR (Fig. 2) were 101.5 TCID50/mL for type 2 PRRSV-infected cells and 102 infected cells/mL for PCV2. Despite using the same primer set (N21/N26 and N22/N24) for the detection of type 2 PRRSV, the method developed in this study was less sensitive (101.5 TCID50/mL) than that of a previously published nested PCR method (101.0 TCID50/mL) [9]. The probable reason for this difference is that our method was a multiple-template PCR reaction (containing templates of both type 2 PRRSV and PCV2 in a single tube), whereas the previous method [9] was a single-template PCR reaction (containing a template of either type 1 or type 2 PRRSV). The simultaneous presence of different templates might produce competition for components of the PCR mixture, and the amounts of amplified products might be reduced. When compared to published RT-nested PCR for PRRSV [9] and PCR for PCV2 [13], the results of our duplex nested RT-PCR (Table 1) were incongruent for only two samples. The kappa statistic, which measures agreement between methods, was 0.867, indicating a good level of agreement, one that is beyond that from chance.
With increased demands for PRRSV and PCV2 detection in wasting or dead pigs, PCR-based methods have been commonly adopted. The present PCR-based study relied on the fact that genomic DNA of PCV2 has to be transcribed into functional RNAs during replication. It thus simplified the nucleotide extraction step for detection of both the RNA and DNA of the genome. The other advantage was that the duplex nested RT-PCR allowed simultaneous detection of type 2 PRRSV and PCV2 in one tube. Our method also included an advantage from a previous study (i.e., designing primers on the relatively genetically stable regions: ORF6 and ORF7) to overcome the genetic variation within each PRRSV genotype [9]. As shown in Fig. 3, the majority of sequences at the primer binding sites (boxes) matched the sequences of each primer. The results imply that the type 2-PRRSV-specific primers used in the duplex nested RT-PCR have the potential to overcome the genetic heterogeneity of type 2 PRRSV. As a result, the method developed in this study could be broadly adopted by other agencies or researchers. Although, hot-start multiplex PCR for classical swine fever virus, African swine fever virus, PCV2, PRRSV, and porcine parvovirus with nucleic acid containing both DNA and RNA, as in the Qiagen DNA/RNA extraction kit, was already developed [4], the RNA-based duplex nested PCR approach described in this study is a convenient, alternative way to detect type 2 PRRSV and PCV2 simultaneously.
Figures and Tables
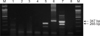 | Fig. 1Cross-reactivity of the duplex nested reverse transcriptase polymerase chain reaction method. Specific fragments of 347 bp (type 2 porcine reproductive and respiratory syndrome virus [PRRSV]) and 265 bp (porcine circovirus type 2 [PCV2]) were amplified with this method. Shown are a 100 bp ladder (lane M), PCV1-infected PK15 cells of different lots (lanes 1–3), PCV1-free PK15 cells (lane 4), PCV2-infected PK15 cells (lane 5), type 2 PRRSV (lane 6), PRRSV and PCV2 co-infected tissue (lane 7), and MARC-145 cells (lane 8). |
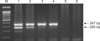 | Fig. 2Detection limits of the duplex nested reverse transcriptase polymerase chain reaction (RT-PCR) method. A mixture of 105 porcine circovirus type 2 (PCV2)-infected PK15 cells/mL and 105.5 TCID50/mL type 2 porcine reproductive and respiratory syndrome virus (PRRSV) was 10-fold diluted. The limited detection of duplex nested RT-PCR was 101.5TCID50/mL for type 2 PRRSV and 102 infected cells/mL for PCV2. A 100 bp DNA ladder (lane M), along with stock of mixture (lane 1) and serial 10-fold dilutions of the stock from 10−1 to 10−5 (lanes 2–6). |
 | Fig. 3Alignment of binding sites for primers N21 (A), N26 (B), N22 (C), and N24 (D). Shaded areas indicate the first 5 nucleotides of the 3' end of each primer. The fractions preceding each primer indicates the number of particular sequences out of the 461 genomic sequences of type 2 porcine reproductive and respiratory syndrome virus (PRRSV) (collected in Europe, Asia, and North American) deposited in Genbank to date. For each primer, the majority of sequences at the primer binding sites (boxes) matched with the sequence of primer. |
Acknowledgments
This study was supported by the BioGreen 21 Program, Rural Development Administration (grant No. PJ011184), and by the Bio-industry Technology Development Program (grant No. 114055031SB010), Ministry of Agriculture, Food and Rural Affairs, Korea. This work was also supported by a grant from the KRIBB Initiative program (KGM3121322). The authors would like to thank the Ministry of Science and Technology (project No. KC.04.22/11-15), Socialist Republic of Vietnam.
References
1. Allan GM, McNeilly F, Ellis J, Krakowka S, Meehan B, McNair I, Walker I, Kennedy S. Experimental infection of colostrum deprived piglets with porcine circovirus 2 (PCV2) and porcine reproductive and respiratory syndrome virus (PRRSV) potentiates PCV2 replication. Arch Virol. 2000; 145:2421–2429.

2. Ellis J, Hassard L, Clark E, Harding J, Allan G, Willson P, Strokappe J, Martin K, McNeilly F, Meehan B, Todd D, Haines D. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can Vet J. 1998; 39:44–51.
3. Ellis J, Krakowka S, Lairmore M, Haines D, Bratanich A, Clark E, Allan G, Konoby C, Hassard L, Meehan B, Martin K, Harding J, Kennedy S, McNeilly F. Reproduction of lesions of postweaning multisystemic wasting syndrome in gnotobiotic piglets. J Vet Diagn Invest. 1999; 11:3–14.

4. Giammarioli M, Pellegrini C, Casciari C, De Mia GM. Development of a novel hot-start multiplex PCR for simultaneous detection of classical swine fever virus, African swine fever virus, porcine circovirus type 2, porcine reproductive and respiratory syndrome virus and porcine parvovirus. Vet Res Commun. 2008; 32:255–262.

5. Halbur PG, Paul PS, Frey ML, Landgraf J, Eernisse K, Meng XJ, Lum MA, Andrews JJ, Rathje JA. Comparison of the pathogenicity of 2 US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol. 1995; 32:648–660.

6. Harms PA, Sorden SD, Halbur PG, Bolin SR, Lager KM, Morozov I, Paul PS. Experimental reproduction of severe disease in CD/CD pigs concurrently infected with type 2 porcine circovirus and porcine reproductive and respiratory syndrome virus. Vet Pathol. 2001; 38:528–539.

7. Kennedy S, Moffett D, McNeilly F, Meehan B, Ellis J, Krakowka S, Allan GM. Reproduction of lesions of postweaning multisystemic wasting syndrome by infection of conventional pigs with porcine circovirus type 2 alone or in combination with porcine parvovirus. J Comp Pathol. 2000; 122:9–24.

8. Kim J, Chung HK, Chae C. Association of porcine circovirus 2 with porcine respiratory disease complex. Vet J. 2003; 166:251–256.

9. Kono Y, Kanno T, Shimizu M, Yamada S, Ohashi S, Nakamine M, Shirai J. Nested PCR for detection and typing of porcine reproductive and respiratory syndrome (PRRS) virus in pigs. J Vet Med Sci. 1996; 58:941–946.

10. Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis A, Martelli GP, Mayo MA, Summers MD. Virus Taxonomy. New York: Springer;1995.
11. Pogranichniy RM, Yoon KJ, Harms PA, Sorden SD, Daniels M. Case-control study on the association of porcine circovirus type 2 and other swine viral pathogens with postweaning multisystemic wasting syndrome. J Vet Diagn Invest. 2002; 14:449–456.

12. Rossow KD, Benfield DA, Goyal SM, Nelson EA, Christopher-Hennings J, Collins JE. Chronological immunohistochemical detection and localization of porcine reproductive and respiratory syndrome virus in gnotobiotic pigs. Vet Pathol. 1996; 33:551–556.

13. Yang JS, Song DS, Kim SY, Lyoo KS, Park BK. Detection of porcine circovirus type 2 in feces of pigs with or without enteric disease by polymerase chain reaction. J Vet Diagn Invest. 2003; 15:369–373.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download