Abstract
Enterococcus spp. are normally present in the gastrointestinal tracts of animals and humans, but can cause opportunistic infections that can be transmitted to other animals or humans with integrated antibiotic resistance. To investigate if this is a potential risk in military working dogs (MWDs), we analyzed antibiotic resistance patterns and genetic relatedness of Enterococcus spp. isolated from fecal samples of MWDs of four different age groups. Isolation rates of Enterococcus spp., Enterococcus (E.) faecalis, and E. faecium, were 87.7% (57/65), 59.6% (34/57), and 56.1% (32/57), respectively, as determined by bacterial culture and multiplex PCR. The isolation rate of E. faecalis gradually decreased with age (puppy, 100%; adolescent, 91.7%; adult, 36.4%; and senior, 14.3%). Rates of resistance to the antibiotics ciprofloxacin, gentamicin, streptomycin, sulfamethoxazole/trimethoprim, imipenem, and kanamycin among Enterococcus spp. increased in adolescents and adults and decreased in senior dogs, with some isolates having three different antibiotic resistance patterns. There were indistinguishable pulsed-field gel electrophoresis patterns among the age groups. The results suggest that Enterococcus is horizontally transferred, regardless of age. As such, periodic surveillance studies should be undertaken to monitor changes in antibiotic resistance, which may necessitate modification of antibiotic regimens to manage antibiotic resistance transmission.
Enterococcus spp. are normally present in the gastrointestinal tracts of animals and humans. Enterococcus (E.) faecalis and E. faecium are among the predominant species [25] and can cause opportunistic infections such as urinary tract and nosocomial infections, bacteremia, and endocarditis [5] in humans. Several factors, including virulence genes and biofilm formation, can increase the pathogenicity of Enterococcus spp. The ability of Enterococcus spp. to acquire antibiotic resistance and transfer it to other bacterial species makes it difficult to control and treat enterococcal infections [1].
Enterococci are considered to have relatively high antibiotic resistance, and it has been reported in livestock as well as companion animals that Enterococci can be transmitted to humans via direct contact [2,31415]. In terms of public health and veterinary medicine, it is important to prevent the transmission of multidrug-resistant (MDR) Enterococcus strains between animals or from animals to humans [18202428]. Previous study has observed that animals that were treated with antibiotics in an intensive care unit were a source or zoonotic transmitter of MDR Enterococcus [12].
Military working dogs (MWDs), trained in army-based training centers, live in close contact with their handlers and other soldiers on army bases, thereby increasing the possibility of horizontal transmission of zoonotic pathogens between animals or from animals to their handlers. It is, therefore, necessary to investigate the characteristics of Enterococcus spp. and the possible risk of antibiotic resistance transmission. Enterococcus spp. are a useful indicator of host antibiotic resistance. To date, no studies have investigated the characteristics and genetic relatedness of Enterococcus spp. from large breeds of dogs, such as those that serve as MWDs.
The aims of this study were to investigate the antibiotic resistance of E. faecalis and E. faecium isolates from the feces of MWDs and compare antibiotic resistance rates among four different MWD age groups. In addition, molecular epidemiological analysis using pulsed-field gel electrophoresis (PFGE) was carried out to determine the genetic relatedness of isolates from MWDs and to elucidate the possibility of transmission of antibiotic resistance among dogs within the training center.
Fecal samples of MWDs (n = 65) from a military working dog training center located in Chuncheon, Korea were collected in September and October 2015. The medical history of each dog was obtained through a questionnaire. The breeds were Belgian Malinois (BGM; n = 44), German Shepherd (GS; n = 20), and mixed (BGM/GS; n = 1). Dogs were categorized into four groups according to age: 3–6 weeks (puppy; n = 12), 9–28 weeks (adolescent; n = 13), 2–6 years (adult; n = 27), and ≥ 9 years (senior; n = 13), including five dogs with diseases. The BGM group included all age groups but the GS group only comprised adult and senior dogs; younger GS dogs were not available.
Fresh fecal samples were collected by using sterile devices from dogs' individual cages, except in the case of puppies, for which samples were obtained by rectal swab. The samples were transported in an ice box to the laboratory and processed within 6 h.
One gram of a fecal sample was homogenized with 9 mL Enterococcosel Broth (BD Biosciences, USA). After incubation for 16 to 18 h at 37℃, samples were streaked onto Enterococcosel Agar (BD Biosciences) and incubated at 37℃ for 16 to 18 h. Three to six suspected colonies were selected from each plate, and, to confirm the identity of Enterococcus spp., multiplex PCR was performed as previously described [1116]. Briefly, DNA was prepared by resuspending a single colony in 200 µL InstaGene Matrix (Bio-Rad Laboratories, USA), followed by incubating in a 56℃ water bath for 30 min, and then boiling for 8 min. The solution was the incubated for 3 min on ice and centrifuged at 13,000 × g for 3 min; the supernatant was used as the template for multiplex PCR amplification. E. faecalis ATCC 29212 and E. faecium ATCC 19434 (American Type Culture Collection, USA) were used as positive controls and nuclease-free distilled water as the negative control.
Antimicrobial susceptibility was assessed by the disk diffusion method according to Clinical and Laboratory Standards Institute (CLSI) guidelines [89]. For 34 E. faecalis and 32 E. faecium isolates, 14 antibiotics were used to test susceptibility: ampicillin (AM), 10 µg; amoxicillin-clavulanic acid (AMC), 30 µg; chloramphenicol (C), 30 µg; tetracycline (TE), 30 µg; ciprofloxacin (CIP), 5 µg; erythromycin (E), 15 µg; imipenem (IPM), 10 µg; sulfamethoxazole/trimethoprim (SXT), 23.75 µg/1.25 µg; gentamycin (GM), 10 µg; streptomycin (S), 10 µg; high-level gentamicin (HLGM), 120 µg; high-level streptomycin (HLS), 300 µg; kanamycin (K), 30 µg; and quinupristin/dalfopristin (SYN), 15 µg (BD Biosciences). Each cultured single isolate was suspended in 0.85% NaCl and turbidity was adjusted to 0.5 McFarland. Five antibiotic disks were placed on each Mueller-Hinton agar (BD Biosciences) plate followed by incubation at 37℃ for 24 h. The diameters of the inhibitory zone of the disks were measured and tested isolates were determined to be susceptible, intermediate, or resistant based on the CLSI M100-S24 E. faecalis and E. faecium breakpoint guidelines. Interpretive criteria were adapted from Enterobacteriaceae guidelines as published in CLSI M100-S23 when the breakpoint was unavailable. S. aureus ATCC 25923 and E. faecalis ATCC 29212 were used as controls.
PFGE was carried out based on the Center for Disease Control and Prevention (CDC)'s Pulse Net protocol for Listeria monocytogenes [6]. We modified the procedures according to the CDC's Gram-positive bacteria PFGE protocol. Briefly, Enterococcus isolates were suspended in TE buffer, adjusted to McFarland scale 4.0, and then the suspension was embedded in 1.0% agarose plugs. After lysis of cells in the agarose plugs and the washing steps, plug slices were digested with 20 U of SmaI (Takara Bio, Japan) at 25℃ for 2 h. Electrophoresis was carried out at 6.0 V for 19 h with a ramped pulse time of 1 to 20 sec in 0.5× Tris-Borate-EDTA buffer at 14℃ using CHEF Mapper (Bio-Rad Laboratories). Gels were visualized by using a Gel Doc XR imager (Bio-Rad). Salmonella Braenderup ATCC BAA664 was used as the size marker.
PFGE band patterns were analyzed with BioNumerics software (ver. 6.6; Applied Maths, USA). PFGE dendrograms were generated for cluster analysis based on the Dice similarity coefficient by using the unweighted pair group method with arithmetic mean method. Isolates showing ≥ 90% similarity were grouped as the same PFGE type.
A total of 66 Enterococcus spp. (34 E. faecalis and 32 E. faecium) were isolated from 65 MWD. The isolation rates of Enterococcus spp., E. faecalis, and E. faecium were 87.7% (57/65), 59.6% (34/57), and 56.1% (32/57), respectively; nine samples contained both E. faecalis and E. faecium (Table 1). The isolation rate in E. faecalis decreased (p< 0.05) with age (puppy, 100%; adolescent, 91.7%; adult, 36.4%; and senior, 14.3%), whereas that of E. faecium increased with age (puppy, 8.3%; adolescent, 58.3%; adult, 72.7%; senior, 85.7%). There were no differences in isolation rates between sexes.
Antibiotic resistance rates were determined for 62 isolates from 60 healthy MWDs (Fig. 1). More than 50% of isolates showed resistance to tetracycline, ciprofloxacin, streptomycin, and sulfamethoxazole/trimethoprim. Resistance to ampicillin, imipenem, and high-level streptomycin was detected only in E. faecium. Rates of resistance to tetracycline, ciprofloxacin, and kanamycin were more than 2-fold higher in E. faecium than in E. faecalis isolates. In contrast, E. faecalis isolates exhibited higher resistance to quinupristin/dalfopristin. All isolates were susceptible to chloramphenicol, amoxicillin-clavulanic acid, and high-level gentamicin.
Antibiotic resistance among isolates from different age groups is summarized in Table 2. Resistance rates as well as the number of antibiotics toward which resistance was observed increased from puppies to adult dogs, but decreased in senior dogs (Fig. 2). Resistance to quinupristin/dalfopristin decreased with age, whereas resistance to ciprofloxacin, gentamycin, streptomycin, sulfamethoxazole/trimethoprim, imipenem, and kanamycin first increased with age before decreasing in the senior group. Resistance to erythromycin increased with age, but the rates remained at a low level. Ampicillin, chloramphenicol, amoxicillin-clavulanic acid, high-level gentamicin, and high-level streptomycin were excluded since resistance to these antibiotics was at a low level and was similar across age groups.
MDR patterns were examined in 66 isolates from 65 MWDs (Table 3). A total of 65 isolates (98.5%) were resistant to more than one antibiotic; that is, they exhibited MDR. Four isolates showed resistance to tetracycline, ciprofloxacin, streptomycin, kanamycin, three to ciprofloxacin, sulfamethoxazole/trimethoprim, kanamycin, and three to tetracycline, ciprofloxacin, gentamycin, streptomycin, sulfamethoxazole/trimethoprim, kanamycin. One isolate obtained from a MWD that had diarrhea showed resistance to 11 types of antibiotics.
The genetic relatedness of 34 E. faecalis and 32 E. faecium isolates from 65 MWDs was determined by using PFGE. For E. faecalis, 13–18 restriction fragments obtained by performing SmaI digestion corresponded to 16 different PFGE types (panel A in Fig. 3). Four of the 16 PFGE types were shared by at least two isolates. For E. faecium, 16–24 restriction fragments obtained by PFGE corresponded to 21 different PFGE types (panel B in Fig. 3). Seven of 21 PFGE types were shared by at least two isolates, whereas 14 PFGE types were observed in one isolate. The number of isolates in each PFGE type ranged from one to 11. Most clusters showed indistinguishable patterns among the age groups. Six of 11 boxed clusters within two PFGE results were related to isolates from breeding dogs, puppies, and adolescents living in a building for delivery dogs.
Enterococcus spp. exist as normal gut flora and are constantly exposed to antibiotics; as such, they are widely used as a model for the acquisition of antibiotic resistance in the host [22]. The prevalence and patterns of antibiotic resistance of Enterococcus spp. isolated from livestock and companion animals have been previously described [41026]. Enterococci in animal and human intestine can transfer antibiotic resistance genes to other host commensals [27]. Most previous studies focused on transmission from livestock to humans or on companion animals in relation to veterinary clinics [713], or they investigated the prevalence of E. faecalis and E. faecium in healthy dogs [1719]. However, ours is the first study to examine the characteristics of Enterococcus spp. in MWDs with respect to their living environment and age. In contrast to a previous study that found a higher prevalence of E. faecalis among dogs without prior exposure to antibiotics and a higher prevalence of E. faecium in those that had been previously exposed [19], we found similar prevalence of E. faecalis (59.6%) and E. faecium (56.1%) among isolates. We also observed that the E. faecalis isolation rate decreased with age, whereas that of E. faecium increased with age. This may have been due to the increase in antibiotic usage with age. Additional studies are required in order to clarify the reason for this difference in prevalence of the two species over the course of the animals' life span.
E. faecalis showed higher resistance to quinupristin/dalfopristin than that of E. faecium, which is in accordance with the known sensitivity of each species to quinupristin/dalfopristin [21]. Enterococcus has intrinsic resistance to β-lactam compounds [21]; however, the MWDs in the present study did not show high resistance to ampicillin. Consistent with the frequent use of fluoroquinolones (e.g., enrofloxacin) at MWD veterinary hospitals, we observed a 50% resistance rate to ciprofloxacin among isolates. Another report detected high resistance to kanamycin (42.6%), streptomycin (35.1%), and erythromycin (33.0%) among dogs in Japan [19]. The differences between those results and ours may be attributed to differences in antibiotic usage at the MWD hospital.
We observed three antibiotic resistance patterns with respect to age: quinupristin/dalfopristin resistance decreased uniformly with age; resistance to ciprofloxacin, gentamycin, streptomycin, sulfamethoxazole/trimethoprim, imipenem, and kanamycin increased in adolescence and adulthood and then decreased in senior dogs; and resistance to ampicillin, chloramphenicol, amoxicillin-clavulanic acid, imipenem, high-level gentamicin, and high-level streptomycin remained similar across age groups. In the case of quinupristin/dalfopristin, the constant rate of decrease corresponded to the reduced the isolation rate of E. faecalis. Puppies in this study had not been exposed to antibiotics and, therefore, did not show resistance to ampicillin, erythromycin, chloramphenicol, gentamicin, streptomycin, amoxicillin-clavulanic acid, or imipenem. However, as they age they undergo many kinds of training and activities that could lead to injury and surgery, which would require the administration of antibiotics. In contrast, senior dogs participate in fewer activities and training, and therefore the chances of experiencing trauma and antibiotic usage would be decreased. The resistance to certain types of antibiotic (tetracycline, ciprofloxacin, sulfamethoxazole/trimethoprim, kanamycin, and quinupristin/dalfopristin) detected in puppies may have been transmitted to them from their parents; this is supported by the finding that Staphylococcus aureus present on parental skin can be transferred to infant gut [23]. Thus, in MWDs living together in the same environment, resistance to some types of antibiotic, but not others, changes with age. This implies the need for taking into account the characteristics of resistance according to age when prescribing antibiotics to MWDs.
Our observation that the MDR rates of isolates (98.5%) were much higher than those previously reported in dogs (50.6%) [20] may be explained by the close contact between MWDs in their unique living environment. The resistance to 11 different antibiotics observed in one isolate may have arisen from a long history of exposure over the course of treatments for chronic diarrhea and melena.
PFGE analysis of 34 E. faecalis revealed four different PFGE types (clusters 1, 4, 5, and 13) that had indistinguishable band profiles (90% similarity). Many breeding dogs, puppies, and adolescent dogs that were recently discharged from the building for delivery dogs showed near-identical PFGE patterns due to being in close contact with each other over an extended period. Indeed, seven of 11 isolates from puppies of the same litter showed the same antibiotic resistant pattern (cluster 4). This is in accordance with another study of antibiotic resistance in Enterococcus isolates from dogs and cats in the USA [23] and may be attributed to the fact that the resistance pattern reflects antibiotic resistance genes that were transferred between bacterial species, while PFGE provides an analysis of total DNA and not of specific genes.
The analysis of the 32 E. faecium isolates revealed seven different clusters (clusters 1, 3, 7, 11, 13, 14, and 15) within a wider range of ages than that of E. faecalis. These clusters included those related to the building for delivery dogs (clusters 11 and 13), suggesting possible transmission between adults (≥ 2 years of age), which is likely the result of direct contact with feces during group training. This raises the possibility of transmission of Enterococcus spp. between MWDs by direct contact.
Considering these two PFGE results, most clusters that included at least two isolates had indistinguishable patterns between MWDs in various age groups. This suggests that Enterococcus transfer does not depend on age. Further studies on the genetic relatedness of Enterococcus spp. in MWDs, as well as in their handlers and other soldiers, are needed to assess the risk of this occurrence.
In conclusion, we investigated for the first time, the characteristics of Enterococcus spp. isolates of large-breed dogs from a MWD training center in Korea. The observed changes in rates of resistance to certain antibiotics with age suggest a need to consider the age of dogs before administering antibiotic treatment. We confirmed by PFGE analysis that the building for delivery dogs was a source of antibiotic-resistant strains, but there was also possible transmission between adult dogs outside of this building. Restricting contact between MWDs and immediate disposal of feces in this building and in the training grounds can minimize the risk of horizontal transmission. The results suggest that surveillance studies should be carried out periodically, in order to follow changes in antibiotic resistance of Enterococcus spp. in MWDs, thereby improving the management of antibiotic resistance in MWDs. In addition, molecular epidemiological analyses of MWD handlers and soldiers are required to address the risk of transmission from dogs to humans.
Figures and Tables
 | Fig. 1Antibiotic resistance rates among Enterococcus (E.) faecalis and E. faecium isolated from 60 healthy military working dogs. AM, ampicillin; AMC, amoxicillin-clavulanic acid; C, chloramphenicol; CIP, ciprofloxacin; E, erythromycin; GM, gentamicin; HLGM, high-level gentamicin; HLS, high-level streptomycin; IPM, imipenem; K, kanamycin; S, streptomycin; SXT, sulfamethoxazole/trimethoprim; SYN, quinupristin/dalfopristin; TE, tetracycline. |
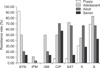 | Fig. 2Age-specific changes in antibiotic resistance rates of Enterococcus spp. isolated from 60 healthy military working dogs. CIP, ciprofloxacin; GM, gentamicin; IPM, imipenem; K, kanamycin; S, streptomycin; SXT, sulfamethoxazole/trimethoprim; SYN, quinupristin/dalfopristin. |
 | Fig. 3Genetic relatedness of Enterococcus (E.) faecalis and E. faecium isolates resistant to antibiotics. (A) E. faecalis (n = 34). (B) E. faecium (n = 32). Band profiles generated by pulsed-field gel electrophoresis are based on 1.5% band matching tolerance. The box indicates the 90% similarity criterion. PFGE patterns were clustered with isolate characteristics including age group, breed, sex, and antibiotic resistance profiles. L and M in sample category indicate E. faecalis and E. faecium isolates, respectively. (a), (b), and (c) indicate siblings. AM, ampicillin; TE, tetracycline; CIP, ciprofloxacin; E, erythromycin; IPM, imipenem; SXT, sulfamethoxazole/trimethoprim; GM, gentamicin; S, streptomycin; HLS, high-level streptomycin; K, kanamycin; SYN, quinupristin/dalfopristin; BGM, Belgian Malinois; GS, German Shepherd. |
Table 3
Antibiotic resistance patterns of Enterococcus spp. isolated from 65 military working dogs

Dogs were categorized into four groups: P, puppy (3–6 weeks); C, adolescent (9–28 weeks); A, adults (1–6 years); S, senior (≥ 9 years). AM, ampicillin; CIP, ciprofloxacin; E, erythromycin; GM, gentamicin; HLS, high-level streptomycin; IPM, imipenem; K, kanamycin; S, streptomycin; SXT, sulfamethoxazole/trimethoprim; SYN, quinupristin/dalfopristin; TE, tetracycline.
Acknowledgments
This work was supported by a National Research Foundation of Korea Grant funded by the Korean Government (NRF-2015R1A1A1A05000990).
References
1. Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012; 10:266–278.

2. Bortolaia V, Guardabassi L. Zoonotic transmission of antimicrobial resistant enterococci: a threat to public health or an overemphasised risk?. In : Sing A, editor. Zoonoses—Infections Affecting Humans and Animals. Focus on Public Health Aspects. New York: Springer;2015. p. 407–431.
3. Buma R, Maeda T, Kamei M, Kourai H. Pathogenic bacteria carried by companion animals and their susceptibility to antibacterial agents. Biocontrol Sci. 2006; 11:1–9.

4. Butaye P, Devriese LA, Haesebrouck F. Differences in antibiotic resistance patterns of Enterococcus faecalis and Enterococcus faecium strains isolated from farm and pet animals. Antimicrob Agents Chemother. 2001; 45:1374–1378.

5. Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR, Harwood VJ. Enterococci in the environment. Microbiol Mol Biol Rev. 2012; 76:685–706.

6. Centers for Disease Control and Prevention (US). Standard operating procedure for PulseNet PFGE of Listeria monocytogenes. Atlanta: 2013.
7. Chung YS, Kwon KH, Shin S, Kim JH, Park YH, Yoon JW. Characterization of veterinary hospital-associated isolates of Enterococcus species in Korea. J Microbiol Biotechnol. 2014; 24:386–393.

8. CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI document M100-S24. Wayne: Clinical and Laboratory Standards Institute;2014.
9. CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. CLSI document M100-S23. Wayne: Clinical and Laboratory Standards Institute;2013.
10. Damborg P, Sørensen AH, Guardabassi L. Monitoring of antimicrobial resistance in healthy dogs: first report of canine ampicillin-resistant Enterococcus faecium clonal complex 17. Vet Microbiol. 2008; 132:190–196.

11. Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995; 33:24–27.

12. Ghosh A, Dowd SE, Zurek L. Dogs leaving the ICU carry a very large multi-drug resistant enterococcal population with capacity for biofilm formation and horizontal gene transfer. PLoS One. 2011; 6:e22451.

13. Ghosh A, Kukanich K, Brown CE, Zurek L. Resident cats in small animal veterinary hospitals carry multi-drug resistant enterococci and are likely involved in cross-contamination of the hospital environment. Front Microbiol. 2012; 3:62.

14. Guardabassi L, Schwarz S, Lloyd DH. Pet animals as reservoirs of antimicrobial-resistant bacteria. J Antimicrob Chemother. 2004; 54:321–332.
15. Hammerum AM, Lester CH, Heuer OE. Antimicrobial-resistant enterococci in animals and meat: a human health hazard? Foodborne Pathog Dis. 2010; 7:1137–1146.

16. Jackson CR, Fedorka-Cray PJ, Barrett JB. Use of a genus- and species-specific multiplex PCR for identification of enterococci. J Clin Microbiol. 2004; 42:3558–3565.

17. Jackson CR, Fedorka-Cray PJ, Davis JA, Barrett JB, Brousse JH, Gustafson J, Kucher M. Mechanisms of antimicrobial resistance and genetic relatedness among enterococci isolated from dogs and cats in the United States. J Appl Microbiol. 2010; 108:2171–2179.

18. Jackson CR, Fedorka-Cray PJ, Davis JA, Barrett JB, Frye JG. Prevalence, species distribution and antimicrobial resistance of enterococci isolated from dogs and cats in the United States. J Appl Microbiol. 2009; 107:1269–1278.

19. Kataoka Y, Ito C, Kawashima A, Ishii M, Yamashiro S, Harada K, Ochi H, Sawada T. Identification and antimicrobial susceptibility of enterococci isolated from dogs and cats subjected to differing antibiotic pressures. J Vet Med Sci. 2013; 75:749–753.

20. Kataoka Y, Umino Y, Ochi H, Harada K, Sawada T. Antimicrobial susceptibility of enterococcal species isolated from antibiotic-treated dogs and cats. J Vet Med Sci. 2014; 76:1399–1402.

21. Kristic CJ, Rice LB, Arias CA. Enterococcal infectiontreatment and antibiotic resistance. In : Gilmore MS, Clewell BD, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Boston: Massachusetts Eye and Ear Infirmary;2014. p. 87–134.
22. Levin BR, Lipsitch M, Perrot V, Schrag S, Antia R, Simonsen L, Walker NM, Stewart FM. The population genetics of antibiotic resistance. Clin Infect Dis. 1997; 24:Suppl 1. S9–S16.

23. Lindberg E, Adlerberth I, Hesselmar B, Saalman R, Strannegård IL, Åberg N, Wold AE. High rate of transfer of Staphylococcus aureus from parental skin to infant gut flora. J Clin Microbiol. 2004; 42:530–534.

24. Lloyd DH. Reservoirs of antimicrobial resistance in pet animals. Clin Infect Dis. 2007; 245:Suppl 2. S148–S152.

25. Moellering RC Jr. Emergence of Enterococcus as a significant pathogen. Clin Infect Dis. 1992; 14:1173–1176.
26. Park JH. A surveillance study of antibiotic resistance of Enterococcus spp. isolated from cattle in Busan area. Annu Rep Busan Metrop City Inst Health Environ. 2013; 23:290–300.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download