Abstract
Bovine adenovirus type 3 (BAdV3) is being used in the development of potential vehicles for gene therapy and vectored vaccine. To that end, a more comprehensive description of BAdV3 biology is essential. In this study, we focused on the role of pIX in BAdV3 virion rescue after full-length BAdV3 genome transfection. Initially, pIX deletion or initiation codon mutation abolished the production of progeny virions, which suggested that pIX was essential for the rescue of BAdV3 containing a full-length genome. Moreover, through transfection of a panel of pIX mutant BAdV3 genomes, we observed that the conserved N-terminus and the putative leucine zipper element (PLZP) were essential for virion rescue, whereas the C-terminus following the coiled-coil domain was non-essential. In addition, swap of the PLZP element and its following region of BAdV3 pIX to corresponding domains of human adenovirus type 5 (HAdV5) did not affect virion production, whereas swap of the entire pIX abolished production of progeny virions. We suggest that failure of the full-length BAdV3 pIX swap might be due to species specificity of its N-terminus region before the PLZP element.
Adenoviruses (AdV) are double-stranded DNA viruses that have been used to develop vectors for gene therapy. The capsid of AdV consists of eight different proteins, including three major proteins (hexon, penton, and fiber) and five minor proteins (VI, VIII, IX, IIIa, and IVa2). Protein IX (pIX), one of the minor capsid proteins, is strongly conserved in Mastadenoviruses and, recently, has received considerable attention [14]. Available information on the structure and function of pIX has been mainly generated by analyzing pIX encoded by human adenovirus (HAdV) types 2 and 5. Each AdV virion contains 240 molecules of pIX [925], which helps to enhance the thermostability of mature virions [5]. Moreover, pIX induces the formation of intranuclear inclusions through sequential relocation of host promyelocytic leukemia (PML) protein into clear amorphous inclusions [19]. Early studies showed that vectors with smaller genomes are inefficiently packaged, whereas vectors with genomes greater than or equal to 27.7 kb are packaged with acceptable efficiencies [15], and pIX is required for packaging genomes with sizes greater than 97% of wild-type (wt) AdV [10]. A recent report suggests that pIX interacts with kinesin-1 through light chain Klc1/2, leading to capsid dismantling [24].
AdV pIX is not essential for formation of progeny virions [5]. Moreover, pIX dependent transactivation of AdV promoters in vitro does not appear to be necessary for the activation of AdV promoters during AdV replication as pIX-deleted AdV grow 2–3-fold less than pIX-containing adenovirus [1321]. Structural and functional analysis of AdV pIX suggests that while the N-terminal domain is essential for incorporation of pIX into the capsid, the C-terminal domain (containing a putative leucine zipper element [PLZP]) is required for self-interaction and presumed trimer formation [26].
Human adenoviruses (HAdV) are one of the most widely used viral vectors for gene therapy and vectored vaccines. However, their clinical use as a gene therapy and in vaccine trials have been hampered by extensive organizational tropism and pre-existing HAdV immunity in the majority of the population. Nonhuman AdV have been developed and proposed as solutions to the problem of pre-existing immune responses. Moreover, nonhuman AdV are potential gene transfer vectors because, although they can enter the human cell, they cannot replicate. In addition, nonhuman AdV vectors have potential uses in animals. In this study, we focused on bovine AdV type 3 (BAdV3). A number of BAdV3 features have been reported, including ease of manipulation of the viral genome, ability to grow to a high titer level, species specificity, ability to overcome the pre-existing immunity against HAdV, as well as better biodistribution and high biosafety, which endow BAdV3 with great potential as a vehicle for vaccine delivery [28]. Moreover, BAdV3 has already been developed as a vector for several vaccines including bovine viral diarrhea virus, bovine herpes virus, and influenza [8]. In order to enhance BAdV3 virion rescue to be as efficient as that from human adenovirus type 5 (HAdV5), we established a novel BAdV3 viral package based on circular BAdV3 genome transfection into VIDO DT1 cells, an engineered cotton rat lung cell line that stably expresses I-SceI endonuclease [6].
The pIX of BAdV3 consists of 125 amino acids and shows 16 to 28% homology with other AdV [17]. BAdV3 pIX protein possesses a transcript that is co-terminal with E1 and encodes a 14 kDa protein detected in virus-infected cells and in mature virions [16]. Moreover, the C-terminus of BAdV3 pIX is exposed on the capsid surface, and that terminus can be used for incorporation of heterologous proteins [27]. In order to investigate the structure and function of BAdV3 pIX during the virus life cycle, a series of recombinant BAdV3 that expressed mutant pIX or chimeric pIX were analyzed.
VIDO DT1 cells were cultivated in minimum essential medium (Gibco, USA) containing 2% fetal bovine serum (FBS; HyClone, USA). The 293A and 293T cells were maintained in Dulbecco's modified Eagle medium (Gibco) supplemented with 10% FBS. Due to similarity of its viral growing characteristics to wtBAdV3 (wtBAdV3), the recombinant BAdV3 virus isolated from genome pFBAV304a.I-SceI, which contains the CMV-enhanced yellow fluorescent protein (EYFP) expression cassette in the E3 region and two I-SceI sites flanking the ITR terminals, was used as wtBAdV3 in this work.
We predicted the structure and function of BAdV3 pIX based on the sequence-to-structure-to-function paradigm and by using the iterative threading assembly refinement (I-TASSER) server [20].
The recombinant plasmids were constructed by applying standard procedures that used restriction enzymes and other DNA-modifying enzymes as directed by the manufacturers. The pFBAV304a.I-SceI was used as template for construction of pIX-deleted BAdV3 genome pFBAV304a.I-SceI-ΔpIX. Subsequently, pFBAV304a.I-SceI-ΔpIX was used as a template for construction of pIX mutant BAdV3 genome plasmids.
As shown in panel A in Fig. 1, a DNA fragment harboring the terminal fragment (nucleotides [nt] 1403 to nt 13578) was isolated from pFBAV304a.I-SceI (containing BAdV3 genome in which the E3 region is replaced with the CMV-EYFP expression cassette, and the ITR terminals are flanked with two I-SceI sites). The fragment was cloned into a BstZ17I-AscI site of 5'-pTKL-3' to create pTK304 [3]. We digested pTK304 with EcoRI and PacI, filled it with T4 DNA polymerase and religated it to eliminate the redundant HpaI site in order to generate pTK304EP (including partial E1 and pIX). Then, we replaced the ORF of pIX (nt 3200 to nt 3574) with the chloramphenicol acetyltransferase (CAT) expression cassette by homologous recombination using HpaI-linearized pTK304EP and a 1040 bp CAT fragment (containing 50 bp homology arms in the 5' and 3' sites and flanked with SbfI enzyme sites) from the template plasmid pTK-bacterial artificial chromosome (BAC) with primers CAT-F and CAT-R [11]. The resulting plasmid was called pTK304EP-CAT.
To construct the shuttle plasmid pTK304-CAT, we isolated a 5385 bp fragment from pTK304EP-CAT with AscI and Eco47III and ligated it into AscI-Eco47III-cleaved pTK304 (panel B in Fig. 1).
The genome pFBAV304a.I-SceI-CAT (in which the ORF of pIX in pFBAV304a.I-SceI was replaced with the CAT expression cassette) was generated by homologous recombination between the AscI-BstZ17I genome fragment from pTK304-CAT and the PmeI-linearized pFBAV304a.I-SceI. The pIX-deleted genome pFBAV304a.I-SceI-ΔpIX was generated by genome pFBAV304a.I-SceI-CAT SbfI digestion and self-ligation (panel C in Fig. 1).
Based on the plasmid pFBAV304a.I-sceI-ΔpIX, a series of pIX-mutated BAdV3 genomes were constructed. The variant mutated pIXs were amplified by polymerase chain reaction (PCR) using primers and template plasmids (Table 1). Finally, the PCR products were inserted into the SbfI site of genome pFBAV304a.I-SceI-mpIX according to the manual for the InFusion HD Cloning Kit (Clontech, USA) (panel D in Fig. 1).
Monolayers of VIDO DT1 cells (2 × 105 cells/well) in 6-well plates were transfected with 5 µg circular plasmid DNAs by using Lipofectin reagent (Invitrogen, USA). After incubation at 37℃ for 7 to 15 days, the transfected cells showing cytopathic effects (CPE) were collected, freeze-thawed five times, and the recombinant viruses serially passaged for three generations to verify whether the recombinant BAdVs were rescued successfully.
To rescue pIX-deleted HAdV type 5 (HAdV5), the resulting plasmid pH5LRedΔpIX was linearized by PacI and transfected into 293A cells together with plasmid pH5R (PacI digested) containing a portion of the HAdV5 genome with a 1878 bp deletion in the E3 region by using Lipofectin reagent (Invitrogen) as per the manufacturer's instructions. Homologous recombination led to the generation of a recombinant virus, named HAdV5Δ pIX. To corroborate the deletion of the pIX region from HAdV5, the viral genome was extracted from 293A cells at 48 h post-infection and sequenced.
VIDO DT1 cells were infected with wt or mutant BAdV3s at a multiplicity of infection (MOI) of 0.1. At indicated times post-infection, the infected VIDO DT1 cells were lysed in the medium by performing five rounds of freeze-thawing, and the amount of virus in each cell lysate was titrated by serial dilution on VIDO DT1 cells, and the median tissue culture infective dose (TCID50) was calculated.
The lysis products of mock- or virus-infected cells were separated by 15% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and transferred to a polyvinylidene fluoride membrane. Nonspecific binding sites on the membrane were blocked overnight with 10% skim milk. After washing twice with TBST (with 0.05% Tween 20), the membranes were incubated with anti-HA monoclonal antibody for 2 h at room temperature, then washed three times with TBST and exposed to alkaline phosphatase (AP)-conjugated goat anti-mouse IgG and developed by using the AP Color Development kit (Bio-Rad Laboratories, USA).
VIDO DT1 cells were placed in a 75 cm2 tissue culture flask. When the cells were approximately 90% confluent, wt or mutant viruses at about 0.1 MOI were added to the flask. After approximately 48 h of infection, the cells were collected in 250 µL medium for virus DNA extraction. Virus DNA was extracted according to the manual of E.Z.N.A. viral DNA Kit (Omega Bio-tek, USA). Subsequently, the viral DNA was identified by digesting with KpnI or SalI and analyzing via agarose gel electrophoresis.
To determine whether pIX is essential for the rescue of BAdV3, the full-length pIX-deleted BAdV3 genome pFBAV304a.I-SceI-ΔpIX was generated by replacing the pIX CDS region with an SbfI enzyme site, as shown in panel A in Fig. 2. The plasmid pFBAV304a.I-SceI-ΔpIX was transfected into VIDO DT1 cells, and the CPE and enhanced EYFP expression were observed at 15 days post-transfection. The recombinant BAdV3 full-length genome expressing pIX (pFBA304a.I-SceI) was used as a control. To compare viral rescue results with that of HAdV5 pIX, the PacI-linearized shuttle plasmid pH5L-DsRed-Mononer or pH5LRedΔpIX was co-transfected into 293A cells with the PacI-linearized genome plasmid pH5R. The CPE and red fluorescent protein (RFP) expression were observed at 15 days post-transfection.
As shown in panel B in Fig. 2, the cells transfected with plasmid pFBAV304a.I-SceI (wtBAdV3), pH5L-DsRed-Mononer (HAdV5-RED, with a similar viral replication feature as wtHAdV5) or pH5LRedΔpIX showed apparent CPE and strong fluorescence (YFP/RFP) at 15 days post-transfection. However, the cells transfected with plasmid pFBAV304a.I-SceI-ΔpIX (BAdV3ΔpIX) showed weak fluorescence, which was absent in subsequent passages, and no CPE were observed (panel C in Fig. 2).
To investigate further whether pIX has a role as a trans-acting factor not as a cis-acting element in the replication of BAdV3 genome, we constructed two plasmids: pFBAV304a.I-SceI-mATGpIX/10GS/HA, in which the initiation codon ATG of BAdV3 pIX was replaced with CTG, and plasmid pFBAV304a. I-SceI-pIX/10GS/HA, in which the pIX/10GS/HA sequence was recombined back in plasmid pFBAV304a.I-SceI-ΔpIX (panel A in Fig. 3). The DNAs of plasmids pFBAV304a.I-SceI-mATGpIX and pFBAV304a.I-SceI-pIX/10GS/HA were individually transfected into VIDO DT1 cells, and the CPE and EYFP expressions were observed. The cells transfected with the DNA of plasmid pFBAV304a.I-SceI DNA (wtBAdV3) or plasmid pFBAV304a.I-SceI-pIX/10GS/HA (BAdV3-pIX/10GS/HA) showed apparent CPE and strong expression of EYFP. However, the cells transfected with plasmid pFBAV304a.I-SceI-mATGpIX/10GS/HA (BAdV3-mATGpIX/10GS/HA) DNA showed very weak EYFP in few transfected cells and no indication of CPE (panel B in Fig. 3). After three passages, the weak EYFP expression was absent (panel C in Fig. 3).
Simultaneously, the expression of recombinant protein was analyzed in plasmid DNAs transfected 293T cells by Western blotting with anti-HA antibody. As seen in panel D in Fig. 3, anti-HA mAb detected a 16 kDa protein in plasmid pFBAV304a.I-SceI-pIX/10GS/HA DNA-transfected cells. However, no such protein was detected in plasmid pFBAV304a.I-SceI-mATGpIX DNA- or plasmid pFBAV304a.I-SceI-ΔpIX DNA-transfected cells.
In panel A in Fig. 4, the amino acid (aa) sequence of BAdV3 pIX was compared with pIX of different HAdV by using the Clustal W program. Although the extent of homology between the pIX of the different AdV was low, two conserved domains were clearly identified in the pIX encoded by bovine and human AdV (panel B in Fig. 4). One of the domains was located at the N-terminus (conserved element, aa 5–41). The other domain was the coiled-coil domain (aa 59–116), which contained a PLZP comprising four leucine and one valine residues, each spaced six residues apart.
To define the functional domains of pIX (panel A in Fig. 5) involved in BAdV3 replication, we constructed several recombinant genome plasmids, as shown in panel B in Fig. 5, (pFBAV304a.I-SceI-pIXΔ1, containing deletion of aa 15–41 of pIX; pFBAV304a.I-SceI-pIXΔ2, containing deletion of aa 76– 104 of pIX; and pFBAV304a.I-SceI-pIXΔ3, containing deletion of aa 117–125 of pIX). VIDO DT1 cells transfected with individual plasmid DNAs were observed for CPE and expression of EYFP. Fifteen days post-transfection, the cells transfected with plasmid pFBAV304a.I-SceI-pIXΔ1 DNA did not exhibit any CPE or EYFP expression (panel C in Fig. 5, BAdV3-pIXΔ1), even after three serial passages (panel D in Fig. 5). The cells transfected with plasmid pFBAV304a.I-SceI-pIXΔ2 showed weak CPE and EYFP expression (panel C in Fig. 5, BAdV3-pIXΔ2), and the CPE and EYFP disappeared after three serial passages (panel D in Fig. 5). In contrast, cells transfected with plasmid pFBAV304a.I-SceI or pFBAV304a.ISceI-pIXΔ3 showed strong CPE and EYFP expression (panel C in Fig. 5, wtBAdV3 or BAdV3-pIXΔ3). The presence of the desired mutation in mutant virus BAdV3-pIXΔ3 was confirmed by performing restriction enzyme analysis of viral DNAs using SalI or KpnI (panel E in Fig. 5).
To determine whether the deletion of aa 117–125 of BAdV3 pIX can affect the replication of BAdV3-pIXΔ3, virus titers were determined by infecting VIDO DT1 cells with freezethawed cell lysates collected from virus-infected cells at indicated times post-infection. As shown in panel F in Fig. 5, mutant BAdV3-pIXΔ3 grew to a similar titer level as that of wtBAdV3. We also measured the thermolability of BAdV3-pIX Δ3; the resulting capsid was as thermostable as the wtBAdV3. These results suggest that the conserved N-terminus (aa 15–41) and the PLZP in the coiled-coil domain (aa 76–104) appear to be essential for BAdV3 rescue in VIDO DT1 cells; in contrast, the C-terminus (aa 117–125) following the coiled-coil domain was non-essential.
To determine the ability of HAdV5 pIX to cis complement BAdV3 genome lacking pIX, we constructed the full-length plasmid pFBAV304a.I-SceI-HpIX in which the pIX gene was replaced with HAdV5 pIX fused to 10 glycine-serine (GS) repeats and an HA tag (panel A in Fig. 6). Initially, transfection of VIDO DT1 cells with plasmid pFBAV304a.I-SceI-HpIX DNA showed CPE and expression of EYFP (panel B in Fig. 6, BAdV3-HpIX). However, the CPE and EYFP expression were limited in distribution to a tiny clump of cells and did not spread to surrounding cells, even after three serial passages (panel C in Fig. 6). These results suggest that HAdV5 pIX could not completely complement the pIX function of BAdV3.
We next examined whether individual domains of BAdV3 pIX could be replaced by homologous domains of pIX encoded by other Mastadenoviruses. We constructed two genome plasmids (panel A in Fig. 6): pFBAV304a.I-SceI-pIXBH1, in which aa 76–125 of BAdV3 pIX were replaced with aa 100–140 of HAdV5 pIX (PLZP element and the C-terminus) and pFBAV304a.I-SceI-pIXBH2, in which aa 105–125 of BAdV3 pIX were replaced with aa 115–140 of HAdV5 pIX (the C-terminus after the coiled-coil domain). Individual pFBAV304a. I-SceI-pIXBH1 and pFBAV304a.I-SceI-pIXBH2 DNAs transfected into VIDO DT1 cells produced CPE and EYFP expression at 15 days post-transfection, and the viruses were named BAdV3-pIXBH1 and BAdV3-pIXBH2, respectively (panel B in Fig. 6). The two chimeric viruses grew well after three passages (panel C in Fig. 6). The results presented in panel D in Fig. 6 show the virion DNA of BAdV3-pIXBH1 and BAdV3-pIXBH2 after further analysis by using KpnI and SalI restriction enzyme, respectively. In addition, the success of the constructed chimeric BAdV3s was further identified by sequencing the chimeric pIX regions.
The expression of chimeric proteins was analyzed by western blotting. As seen in panel E in Fig. 6, the expected molecular weights of the chimeric proteins were detected by anti-HA mAb after chimeric adenovirus transfection into VIDO DT1 cells. Samples of VIDO DT1 cells infected with wtBAdV3 and mock VIDO DT1 cells were used as controls.
To determine if replacement of the homologous domains of BAdV3 pIX with corresponding domains of HAdV5 pIX can affect the replication of the mutant viruses, virus titers were determined. VIDO DT1 cells infected with wt or individual mutant BAdV3s were collected at indicated times postinfection; after freeze-thawing, the cell lysates were analyzed by performing TCID50 assays. As seen in Fig. 6f, mutant BAdV3s grew to a similar titer level as that of wtBAdV3.
HAdV5 polypeptide IX (pIX) is a multifunctional protein involved in inducing nuclear reorganization in infected cells and providing thermostability to mature virions [92324]. pIX stabilizes the AdV capsid structure and functions as a “cement”. The AdV capsids can only take in less than a 35 kb AdV genome (97% of the wt genome) [10]. In this work, HAdV5-RED-ΔpIX has 90.7% of wtHAdV5 genome with deletions in E1A, E1B (replaced by the insertion CMV-DsRed-Monomer), E3, and the gene sequences of pIX, which can grow as efficiently as HAdV5- RED. In an earlier study, we demonstrated that BAdV3 pIX is a 14 kDa protein that has its C-terminus located on the outside of virions and can be used for addition of targeting ligands [27]. Here, we report a detailed analysis of BAdV3 pIX by constructing and analyzing recombinant BAdV3 expressing mutant pIXs.
As the pIXs between BAdV3 and HAdV5 have a wide range of variation in sequences alignment and structure, it is still unknown whether the function of pIX for full-length BAdV3 virion rescue after transfection is similar to that reported for HAdV5. In the present study, the revertant BAdV3-pIX/10GS/HA (wt pIX was replaced by pIX fused with 10GS linker and HA tag) could be successfully rescued. In addition, the genome size of our pIX-deleted (replaced by SbfI enzyme site, 34512 bp) BAdV3 (BAdV3-ΔpIX) is similar to the size of wtBAdV3 genome (34446 bp). The genome of the start codon mutant (ATG to CTG, 34999 bp) BAdV3 (BAdV3-mATGpIX/10GS/HA) is 101.6% of wtBAdV3 genome. Neither BAdV3-ΔpIX nor BAdV3-mATGpIX/10GS/HA were capable of forming infectious virions following full-length viral genome transfection into VIDO DT1 cells. These observations are similar to the results of previous studies on HAdV5 in which the HAdV5 that lacked about 5349 bp and contained an insertion of 5.4 kb or 6 kb in the site of the deletion was almost non-infectious resulting in minute plaques after long incubation periods and exhibited no CPE [10]. These results suggest that pIX, although a minor component of BAdV3, is essential for the packaging of full-length genomes and its function is the same as that of pIX of HAdV5.
Although the position of the pIX protein is conserved in the genomes of Mastadenoviruses, some studies have demonstrated significant variation in the structure, subcellular localization, and function of AdV protein homologs encoded by different Mastadenoviruses [124]. Several pieces of evidence suggest that pIX encoded by members of Mastadenoviruses that infect humans and animals may differ in structure and function. First, although pIX encoded by different HAdV show significant (45%–100%) homology, pIX encoded by BAdV3 showed homologies of 16% to 28% to pIX encoded by HAdV [17]. Second, the alanine-rich central domain identified in pIX encoded by HAdV appear absent in pIX encoded by BAdV3 [18]. Third, the predicted 3-D structures of pIX encoded by BAdV3 and HAdV5 show great difference at the conserved N-terminus.
In HAdV, residues 22 to 28, located in the N-terminal domain of pIX, was essential for pIX incorporation into the capsid and for virion thermostability [18]. Consistent with the HAdV, these residues were conserved among animal AdV serotypes [26]. Our inability to successfully rescue mutant BAdV3-pIXΔ1 suggests that the N-terminus of pIX has an essential role for BAdV3 replication. The atomic models of HAdV5 pIX based on cryo-electron microscopy revealed that the N-terminus was tied by special construction, such as a hydrophobic core, which was created by the conserved amino acid to participate in AdV capsid assembly [12]. We speculate that the deletion of conserved amino acids of pIX from BAdV3 would affect the pIX participating in the capsid, preventing AdV assembly and virus rescue.
Besides the involvement of the conserved N-terminal domain, the coiled-coil domain was involved in the trimer formation of pIX encoded by HAdV5 [7] and was not essential for the incorporation and thermostability of capsid [26]. Previous studies indicated that pIX, especially the PLZP, ensured optimal viral proliferation by inducing sequestration of PML (promyelocytic leukemia) protein, nuclear reorganization and disassembly, and viral genome nuclear access [18192224]. In this work, although the pIX coiled-coil domain (including PLZP) was deleted, BAdV3 (BAdV3-pIXΔ2) was rescued at 15 days post-transfection. However, the mutant BAdV3-pIXΔ2 could not be amplified during successive passages. It is possible that the coiled-coil domain containing a leucine zipper is involved in inactivation of PML bodies, and is required for the efficient replication/proliferation of BAdV3.
The conserved N-terminal domain and the coiled-coil domain (containing a PLZP element) were the only two putative functional domains for BAdV3 observed following sequence alignment. In the present study, we confirmed that the only two putative functional domains of pIX were also the essential domains for BAdV3 replication. To explore whether the conserved N-terminal domain and coiled-coil domain of pIX could be swapped among different species of Mastadenoviruses, mutant BAdV3s expressing chimeric pIX containing replacement of aa 76–125 of BAdV3 pIX with the corresponding region of HAdV5 pIX (PLZP element and C-terminus) (BAdV3-pIXBH1) and replacement of aa 105–125 of BAdV3 pIX with the corresponding region of HAdV5 (C-terminus) (BAdV3-pIXBH2) could be successfully rescued and grew as efficiently as BAdV3 containing wt pIX. A parallel experiment using the corresponding region of PAdV-3 pIX was also successful, confirming that despite absence of typical aa sequence in the C-terminus of PAdV-3 pIX, the predicted secondary structure of this region appears as a helix bundle, which is similar to the C-terminus region of BAdV3 pIX or HAdV5 pIX (coiled-coil domain). These results suggest that the function of the coiled-coil domain of pIX encoded by different Mastadenoviruses depends on the secondary or tertiary structures rather than the primary amino acid sequence.
However, the rescue of mutant BAdV3-HpIX containing a replacement of pIX of BAdV3 with pIX of HAdV5 was not successful. As predicted from their 3-D structures(data not shown), the region from the start of the PLZP element to the end of C-terminus appeared similar in BAdV3 pIX and HAdV5 pIX, while the conserved N-terminus of pIX showed significant differences. We speculate that the failure of BAdV3-HpIX virions rescue might be related to their structural difference at the N-terminus.
Taken together, our results reveal that pIX is essential and acts as a trans-acting factor for full-length BAdV3 viral genome rescue. Furthermore, we show that the conserved N-terminus and the coiled-coil domain of pIX are essential for virions rescue following BAdV3 viral genome transfection. In addition, the conserved N-terminus could not be switched among AdV of different species. No doubt, pIX has additional secrets to be revealed, and those revelations should be taken into consideration when developing BAdV3-based vectors.
Figures and Tables
 | Fig. 1Strategy used for the generation of recombinant BAdV3 plasmids. (A) The flowchart for construction of plasmid pTK304EP-chloramphenicol acetyltransferase (CAT). Firstly, a 12176 AscI-BstZ17I DNA fragment (nucleotide [nt] 1403 to nt 13578) from pFBAV304a.I-SceI was ligated into AscI-BstZ17I- cleaved 5'- pTKL- 3' to create pTK304. Then, digested plasmid pTK304 with EcoRI and PacI was blunted and self-ligated for eliminating the extra HpaI site to generate pTK304EP. Finally, to construct plasmid pTK304EP-CAT gene, a 1040 bp CAT fragment (containing 50 bp homology arms in 5' and 3' sites and flanked with SbfI enzyme sites) was inserted into HpaI-linearized pTKEP304 in Escherichia coil BJ5185 by homology recombination. (B) The flowchart for construction of shuttle plasmid pTK304-CAT. A 5385 bp fragment isolated from pTK304EP-CAT using AscI and Eco47III was ligated into AscI-Eco47III-cleaved pTK304 to created final shuttle plasmid pTK304-CAT. (C) The flowchart for construction of pIX deleted BAdV3 plasmid pFBAV304a. I-SceI-ΔpIX. AscI-BstZ17I-cleaved pTK304-CAT was cloned into PmeI-linearized pFBAV304a.I-SceI by homology recombination in Escherichia coil BJ5185 to construct pFBAV304a.I-SceI-CAT. Then, the pIX deleted recombination plasmid pFBAV304a.I-SceI-ΔpIX was constructed by digesting the recombinant pFBAV304a.I-SceI-CAT with SbfI and religated to release CAT expression cassette. (D) The flowchart for construction of recombinant BAdV3s expressing genes of variant mutated pIXs. The variant pIX mutations were amplified by polymerase chain reaction using primers and templates (Table 1), then inserted in SbfI site of pFBAV304a.I-SceI-ΔpIX using In-Fusion HD Cloning Kit. CAT with 50 bp homology to pIX ORF upstream and downstream and flanked with SbfI sites. The asterisk represents the site for PacI& EcoRI digestion, blunting, and self-ligation. |
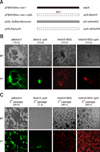 | Fig. 2Generation of pIX-deleted Adenoviruses variants. (A) Schematic representation of the pIX mutants. The pIX-deleted mutants were introduced by homology recombination (pFBAV304a.IsceI-ΔpIX, replace pIX by SbfI enzyme site) and enzyme cleavage (pH5LRedΔpIX, deletion pIX by AflII & MunI digestion and blunt ligation). The full name of genome plasmids are listed on the left and the corresponding pIX mutant proteins expressed by the recombinant viruses were listed on the right. “■” represents the CDS of BAdV3's pIX. “ |
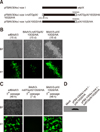 | Fig. 3Generation of start codon mutant and pIX/10GS/HA fusion gene. (A) Schematic representation of the pIX variants. Plasmid names are listed on the left and the corresponding pIX mutant proteins expressed by recombinant viruses are listed on the right. “■” represents the CDS of BAdV3's pIX, “ |
 | Fig. 4Conserved amino acid regions in pIX. (A) Amino acid sequence alignment of pIX from Bovine (b) AdV and several human (h) AdV serotypes (Clustal W). Fully conserved residues are shaded black. Residues that occur in more than 50% of the sequences are shaded gray. Conserved sequence elements are boxed. The symbol “♦” denotes identical amino acid residues. (B) Amino acid sequence of BAdV3 pIX. The conserved sequence element and the coiled-coil domain are underlined, and the square bracket delineates the putative leucine zipper (2-ZIP server) [26,27]. |
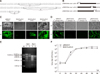 | Fig. 5Generation of pIX truncated BAdV3s (BAdV3-pIXΔ1, BAdV3-pIXΔ2, and BAdV3-pIXΔ3). (A) Sequence of the BAdV3 pIX gene. (B) The schematics of truncated pIX mutants. The amino acid positions above the polyline depict the truncated region of pIX. Deletions of the three conserved domains were introduced by using the In-Fusion HD Cloning Kit. The names of genome plasmids are listed on the left and the corresponding names of the pIX mutant proteins expressed by rescued viruses are listed on the right. (C) The genome plasmids in panel B were transfected into VIDO DT1 cells for viral genome rescue, respectively. At 15 days post-transfection, cytopathic effect (CPE) and enhanced yellow fluorescent protein (EYFP) expression were examined. (D) The VIDO DT1 cells were infected by second passage of wtBAdV3, BAdV3-pIXΔ1, BAdV3-pIXΔ2, or BAdV3-pIXΔ3 virions and the CPE and EYFP expression examined. (E) Viral DNAs were extracted from VIDO DT1cells infected with BAdV3-pIXΔ3 or wtBAdV3. After SalI digestion, a 8216 bp fragment in wtBAdV3 viral DNA (lane 2) was cleaved into two fragments (4690 bp and 3550 bp) in BAdV3-pIXΔ3 viral DNA (lane 1), and post-KpnI digestion, a 6730 bp fragment in wtBAdV3 viral DNA (lane 4) was cleaved into two fragments (3578 bp and 3176 bp) in BAdV3-pIXΔ3 viral DNA (lane 3). Marker (M) sizes are shown in bases. (F) Dynamics of wtBAdV3 and BAdV3-pIXΔ3 growth in VIDO DT1 cells. The virus titers were determined at indicated times by plaque-forming unit (PFU) analysis. BF, bright field; FF, fluorescence field; wt, wild-type. |
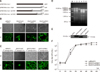 | Fig. 6Generation of chimeric BAdV3s (BAdV3-HpIX, BAdV3-pIXBH1 and BAdV3-pIXBH2). (A) The schematics of chimeric pIXs. “ |
Acknowledgments
This work was supported by grants from the Agriculturerelated Special Fund for 13th Five Year Plan (2016YFD0500306), Saskatchewan Health Research Foundation (SHRF) of the Natural Sciences and Engineering Research Council of Canada, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences (grant No. SKLVBF201405), and the National Natural Science Foundation of China (grant No. 31101803/C1803).
References
1. Bangari DS, Shukla S, Mittal SK. Comparative transduction efficiencies of human and nonhuman adenoviral vectors in human, murine, bovine, and porcine cells in culture. Biochem Biophys Res Commun. 2005; 327:960–966.

2. Blanchette P, Wimmer P, Dallaire F, Cheng CY, Branton PE. Aggresome formation by the adenoviral protein E1B55K is not conserved among adenovirus species and is not required for efficient degradation of nuclear substrates. J Virol. 2013; 87:4872–4881.

3. Cadiñanos J, Bradley A. A Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007; 35:e87.
4. Cheng CY, Gilson T, Wimmer P, Schreiner S, Ketner G, Dobner T, Branton PE, Blanchette P. Role of E1B55K in E4orf6/E1B55K E3 ligase complexes formed by different human adenovirus serotypes. J Virol. 2013; 87:6232–6245.

5. Colby WW, Shenk T. Adenovirus type 5 virions can be assembled in vivo in the absence of detectable polypeptide IX. J Virol. 1981; 39:977–980.

6. Du E, Tikoo SK. Efficient replication and generation of recombinant bovine adenovirus-3 in nonbovine cotton rat lung cells expressing I-SceI endonuclease. J Gene Med. 2010; 12:840–847.

7. Fabry CM, Rosa-Calatrava M, Moriscot C, Ruigrok RW, Boulanger P, Schoehn G. The C-terminal domains of adenovirus serotype 5 protein IX assemble into an antiparallel structure on the facets of the capsid. J Virol. 2009; 83:1135–1139.

8. Ferreira TB, Alves PM, Aunins JG, Carrondo MJ. Use of adenoviral vectors as veterinary vaccines. Gene Ther. 2005; 12(Suppl 1):S73–S83.

9. Furcinitti PS, van Oostrum J, Burnett RM. Adenovirus polypeptide IX revealed as capsid cement by difference images from electron microscopy and crystallography. EMBO J. 1989; 8:3563–3570.

10. Ghosh-Choudhury G, Haj-Ahmad Y, Graham FL. Protein IX, a minor component of the human adenovirus capsid, is essential for the packaging of full length genomes. EMBO J. 1987; 6:1733–1739.

11. Kunec D, Hanson LA, van Haren S, Nieuwenhuizen IF, Burgess SC. An overlapping bacterial artificial chromosome system that generates vectorless progeny for channel catfish herpesvirus. J Virol. 2008; 82:3872–3881.

12. Liu H, Jin L, Koh SBS, Atanasov I, Schein S, Wu L, Zhou ZH. Atomic structure of human adenovirus by cryoEM reveals interactions among protein networks. Science. 2010; 329:1038–1043.

13. Lutz P, Rosa-Calatrava M, Kedinger C. The product of the adenovirus intermediate gene IX is a transcriptional activator. J Virol. 1997; 71:5102–5109.

15. Parks RJ, Graham FL. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packing. J Virol. 1997; 71:3293–3298.

16. Reddy PS, Chen Y, Idamakanti N, Pyne C, Babiuk LA, Tikoo SK. Characterization of early region 1 and pIX of bovine adenovirus-3. Virology. 1999; 253:299–308.

17. Reddy PS, Idamakanti N, Zakhartchouk AN, Baxi MK, Lee JB, Pyne C, Babiuk LA, Tikoo SK. Nucleotide sequence, genome organization, and transcription map of bovine adenovirus type 3. J Virol. 1998; 72:1394–1402.

18. Rosa-Calatrava M, Grave L, Puvion-Dutilleul F, Chatton B, Kedinger C. Functional analysis of adenovirus protein IX identifies domains involved in capsid stability, transcriptional activity, and nuclear reorganization. J Virol. 2001; 75:7131–7141.

19. Rosa-Calatrava M, Puvion-Dutilleul F, Lutz P, Dreyer D, de Thé H, Chatton B, Kedinger C. Adenovirus protein IX sequesters host-cell promyelocytic leukaemia protein and contributes to efficient viral proliferation. EMBO Rep. 2003; 4:969–975.

20. Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010; 5:725–738.

21. Sargent KL, Meulenbroek RA, Parks RJ. Activation of adenoviral gene expression by protein IX is not required for efficient virus replication. J Virol. 2004; 78:5032–5037.

22. Souquere-Besse S, Pichard E, Filhol O, Legrand V, Rosa-Calatrava M, Hovanessian AG, Cochet C, Puvion-Dutilleul F. Adenovirus infection targets the cellular protein kinase CK2 and RNA-activated protein kinase (PKR) into viral inclusions of the cell nucleus. Microsc Res Tech. 2002; 56:465–478.

23. Stewart PL, Fuller SD, Burnett RM. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 1993; 12:2589–2599.

24. Strunze S, Engelke MF, Wang IH, Puntener D, Boucke K, Schleich S, Way M, Schoenenberger P, Burckhardt CJ, Greber UF. Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host Microbe. 2011; 10:210–223.

25. van Oostrum J, Burnett RM. Molecular composition of the adenovirus type 2 virion. J Virol. 1985; 56:439–448.

26. Vellinga J, van den Wollenberg DJ, van der Heijdt S, Rabelink MJ, Hoeben RC. The coiled-coil domain of the adenovirus type 5 protein IX is dispensable for capsid incorporation and thermostability. J Virol. 2005; 79:3206–3210.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 ” represents the CDS of HAdV5's pIX, and “□” represents the deletion of the pIX's CDS of BAdV3 or HAdV5. “SbfI” indicates the use of SbfI enzyme site to replace the pIX's CDS of BAdV3. (B) Recombinant virions were rescued either in VIDO DT1 cells (BAdV3) or 293A cells (HAdV5). The cytopathic effect and fluorescence were observed at 15 days post-transfection. (C) The cells (VIDO DT1 or 293A) were infected by 2nd passage of wtBAdV3 or BAdV3ΔpIX virions, or by 2nd passage of HAdV5-RED or HAdV5-RED-ΔpIX virions. BF, bright field; FF, fluorescence field; wt, wild-type.
” represents the CDS of HAdV5's pIX, and “□” represents the deletion of the pIX's CDS of BAdV3 or HAdV5. “SbfI” indicates the use of SbfI enzyme site to replace the pIX's CDS of BAdV3. (B) Recombinant virions were rescued either in VIDO DT1 cells (BAdV3) or 293A cells (HAdV5). The cytopathic effect and fluorescence were observed at 15 days post-transfection. (C) The cells (VIDO DT1 or 293A) were infected by 2nd passage of wtBAdV3 or BAdV3ΔpIX virions, or by 2nd passage of HAdV5-RED or HAdV5-RED-ΔpIX virions. BF, bright field; FF, fluorescence field; wt, wild-type.
 XML Download
XML Download