Abstract
Well characterized, stable, p16-defective canine mammary cancer (CMT) cell lines and normal canine mammary epithelial cells were used to investigate expression of the major breast cancer-specific hormone receptors estrogen receptor alpha (ER1) and progesterone receptor (PR) as well as luminal epithelial-specific proto-oncogenes encoding c-erbB-1 (epidermal growth factor receptor/EGFr), c-erbB-2/HER2, c-erbB-3, and c-erbB-4 receptors. The investigation developed and validated quantitative reverse transcriptase polymerase chain reaction assays for each transcript to provide rapid assessment of breast cancer phenotypes for canine cancers, based on ER1, PR, and c-erbB-2/HER2 expressions, similar to those in human disease. Roles for relatively underexplored c-erbB-3 and c-erbB-4 receptor expressions in each of these breast cancer phenotypes were also evaluated. Each quantitative assay was validated by assessment of amplicon size and DNA sequencing following amplification. Differential expression of ER1, PR, and c-erbB-2 in CMT cell lines clearly defined distinct human-like breast cancer phenotypes for a selection of CMT-derived cell lines. Expression profiles for EGFr family genes c-erbB-3 and c-erbB-4 in CMT models also provided an enriched classification of canine breast cancer identifying new extended phenotypes beyond the conventional luminal-basal characterization used in human breast cancer.
Canine mammary carcinomas are the most frequent cancer diagnosed in reproductively intact female dogs and represent spontaneous neoplasms of epithelial tissues [11]. Diagnosis and staging of these tumors is currently achieved by using a combination of pathology and physical diagnosis [15], although attempts have been made to apply immunohistochemical analysis to this problem, which can be challenging due to extended/variable fixation and sectioning artifacts and the requirement for specific target labeling [162225]. As a result, diagnosis and staging of canine mammary tumors is still largely based on standard traditional pathological analysis.
In addition to standard pathological analysis, expression analysis arrays are commonly used to evaluate gene expression profiles of human breast tumor biopsies. Genes of interest include estrogen receptor alpha (ERα or ER1), progesterone receptor (PR), and the HER2 receptor tyrosine kinase (human epidermal growth factor receptor 2 or c-erbB-2/Neu), and these expression profiles provide important predictive power with respect to clinical outcome and treatment response [21]. Expressions of these three genes have been used, in part, to define subtypes of human breast cancer including luminal A, luminal B, HER2-positive, and basal/triple negative phenotypes [1232427]. Each of these phenotypes behave, and are treated, differently. Effectiveness of treatment is dependent on a precise diagnosis and appropriate treatment [48].
Spontaneously occurring canine mammary cancers have been used to model human breast cancer in terms of causative mutations as well as for development of new treatment strategies for both species [271820]. A more rapid and quantitative assay, based on quantitative reverse transcriptase polymerase chain reaction (QrtPCR), of canine mammary cancer transcripts would promote more precise mRNA-based phenotype determination of canine breast cancer subtypes, promote utilization of more effective and precise treatments, and promote development or adaptation of new therapies designed to target-specific breast cancer subtypes in both species.
Investigation of canine mammary cancer genetics has been a long-standing focus of our laboratory both for the purpose of understanding genetic defects that promote these tumors and of providing a more comparative perspective on mammary cancers in dog and human patients [261213181920]. Such immune-intact models provide the opportunity for development of advanced immune-modulatory therapies and represent a more similar pathology than murine models [7]. In this report, we describe the successful development of validated QrtPCR assays for six receptor genes designed to specifically promote and extend the application of human breast cancer subtypes to canine mammary cancer. The analysis establishes equivalent evaluations for canine breast cancer, based on expression of estrogen and progesterone receptors, and extends the analysis to all four HER/EGFr genes, including cHER2 (canine HER2). These assays allow identification of new potential phenotypes that should promote development of improved treatments focused more precisely on defined mammary cancer phenotypes in this species.
Canine mammary tumor (CMT) cell lines (Table 1) were originally obtained from Dr. L. Wolfe or were similarly derived by the authors. CMT cell lines and canine mammary epithelial cells (CMECs) were cultured in L-15 medium (Gibco, USA) with antibiotics (Sigma, USA), and 10% FBS (fetal bovine serum; HyClone, USA) in tissue culture flasks (Corning, USA) at 37℃ (95% air; 5% CO2) as previously described [61213182030]. All use of animal cell cultures was under the guidance and review of the Auburn University Institutional Animal Care and Use Committee (protocols PRN 2009-1633, PRN 2007-1155, PRN 2005-0826).
Cell cultures were grown (75–80% confluence), total RNA isolated by using High Pure RNA Isolation kits (Roche Diagnostics, Germany), and RNA concentration determined (absorbance at 260 nm [31]). Transcripts encoding canine estrogen receptor alpha (ER1), progesterone receptor (PR), and epidermal growth factor receptors 1–4 (EGFr/c-erbB-1/HER1, c-erbB-2/HER2, c-erbB-3/HER3, and c-erbB-4/HER4) were amplified by using sequence-specific primers designed based on canine transcript or genomic sequences in Genbank (National Center for Biotechnology Information, USA; Table 2). Synthesis and amplification of cDNA was performed via QrtPCR by using a Bio-Rad iCycler iQ Multicolor PCR Detection System using an ABsoluteTM QPCR SYBR Green Fluorescein Mix (Thermo Scientific, USA) [12].
The first QrtPCR step began with reverse transcription of total RNA (0.5–1 µg) from each cell line being added to a reaction mixture containing a reverse transcription supermix (Bio-Rad Laboratories, USA). Final 20 µL reactions were incubated for 5 min at 25℃, 30 min at 42℃, and 5 min at 85℃. The cDNAs were used as templates for QrtPCR (10 µL reactions containing 50 ng cDNA added to SYBR-Green master mix; Bio-Rad Laboratories). Cycling conditions were initial polymerase activation and denaturation at (95℃, 30 sec) and then 35 cycles of two-step amplification including denaturation at 95℃ for 15 sec and annealing at the respective annealing temperature of each gene (Table 2) for 20 to 30 sec. Amplification was quantitatively assessed at each annealing step. Before QrtPCR assay, RNA quality was validated by performing a replicate assay without no reverse transcription to confirm the absence of genomic DNA contamination (data not shown). Amplification efficiency (90–100%) was determined from the standard curve for each template. Relative normalized expression was calculated using the ΔΔCt (or Cq) method. Quantitative gene expression data were analyzed by using CFX Manager 3.0 software (Bio-Rad Laboratories) based on expression of housekeeping gene transcripts encoding GAPDH and ribosomal protein L37 to provide positive reactions and loading/RNA content and RNA integrity controls [28]. To indicate absolute amounts of amplicons recovered in a semi-quantitative assay before normalization of the data, validation of transcript size was performed by performing agarose gel electrophoresis of amplicons as previously described [13].
Following amplification, amplicons were gel purified, cloned into vector pCR2.1 (Invitrogen, USA) and their identity confirmed by DNA sequencing of several independent clones (Harvard University Genomics and Sequencing Laboratory, USA). Sequence analysis was performed using Vector NTI (Invitrogen). RNA extraction, QrtPCR, and sequencing were performed as previously described [13]. All assays were performed a minimum of three times, standard deviations calculated, and Student's t-tests used to determine significance (p≤0.05).
Well characterized CMT cell lines (CMT9, CMT12, CMT27, CMT28, CMT47, CMT119) and normal CMEC cultures were used as models of breast cancer–derived cells and normal cells derived from breast tissue (Table 1, Fig. 1). Cell lines were used to establish assays and expression profiles for canine estrogen receptor alpha and progesterone receptors as well as for four members of the canine c-erbB receptor family (epidermal growth factor receptor/HER1, HER2/c-erbB-2/Neu, c-erbB-3/HER3, and c-erbB-4/HER4) based on authentic canine genomic sequences. Sequences were selected based on their uniqueness and suitability for QrtPCR and for the ability to promote amplification without cross-priming of closely related gene family members (Table 2). All QrtPCR amplification products were amplified as single bands of the molecular weight predicted when evaluated by agarose gel electrophoresis and all were validated by DNA sequencing to ensure amplicon authenticity. Additionally, all QrtPCR amplifications were normalized to two independent control genes encoding GAPDH and ribosomal protein L37 to ensure no effects of loading, handling or recovery affected results. All results were calculated as relative values compared to levels observed in CMEC for the appropriate gene, which were set to 1 to normalize for relative expression against normal mammary epithelial cells (Figs. 2 and 3).
A selection of six established cell lines, derived from canine mammary carcinomas or adenocarcinomas and representing the best characterized CMT cell lines, as well as several lines exhibiting varied phenotypes in vitro, were analyzed. Express profiles for the estrogen and progesterone receptors were first investigated to determine if CMT cells expressed these receptors at levels comparable to those in CMECs. QrtPCR analysis of the estrogen receptor alpha gene expression revealed that five of the six CMT cell lines expressed only trace levels (panel A in Fig. 2). The exception was CMT12, in which estrogen receptor alpha mRNA expression levels were approximately 45% of levels detected in CMECs. Similarly, QrtPCR analysis of progesterone receptor gene expression revealed that four of six CMT cell lines expressed only trace levels of this receptor's mRNA (panel B in Fig. 2). The exceptions were CMT119, in which progesterone expression was observed at approximately 60% of the levels detected in CMECs, and CMT47 cells, in which progesterone receptor expression was 2.7-fold higher than that in CMECs. In Fig. 2, all differences that were statistically significant (p≤0.05) are noted by an asterisk.
Expression profiles were extended to include four members of the c-erbB gene family. EGFr (c-erbB-1) was expressed at low/moderate levels in CMT cell lines, relative to the levels in CMECs, with EGFr expression in four CMT lines not exceeding 64% of the CMEC levels (panel A in Fig. 3). However, CMT12 cells expressed EGFr at approximately 1.5-fold higher than CMECs and CMT119 cells expressed EGFr at levels approximately 5.5-fold higher than CMECs. In contrast, HER2 was overexpressed in all but one of the CMT cell lines (panel B in Fig. 3). HER2 overexpression ranged from 1.5-fold in CMT28 cells to 6.6-fold in CMT12 cells. CMT9, CMT27, and CMT119 cells exhibited 3.6-fold, 4.8-fold, and 5.6-fold HER2 overexpression, respectively. Only CMT47 cells did not overexpress the HER2 receptor mRNA (approximately 40% of CMEC levels) suggesting a strong bias for HER2 overexpression in most CMT lines.
Because c-erbB-3 and c-erbB-4 genes have been implicated in breast cancer oncogenesis through heterodimer formation with the HER2 receptor, we extended the transcriptional analysis beyond the typical receptors employed in characterization of human breast cancers to include both canine c-erbB-3 and c-erbB-4 transcription profiles. Relative to expression in CMECs, which was very low, expression of c-erbB-3 was low to normal in three of the CMT cell lines but was overexpressed by approximately 8-fold in CMT9 cells, 70-fold in CMT27 cells, and 29-fold in CMT119 cells (Fig. 2C). Relative to expression in CMECs, expression of c-erbB-4 was low/normal in four of the CMT cell lines but overexpressed by 1.3-fold in CMT9 cells, and 2.55-fold in CMT119 cells (Fig. 3D). The data demonstrate that overexpression of c-erbB-3 and c-erbB-4 were detectable and one or both genes were frequently overexpressed in CMT cells.
Correlations between the expressed receptor profiles of common human breast cancer subtypes and those detected in the CMT cells revealed a surprising bias within the six CMT cell lines. Of those CMT cell lines, three were assessed as having a HER2 positive subtype (CMT9, CMT27, and CMT28) due to high levels of HER2 expression and essentially non-detectable levels of estrogen receptor and progesterone receptor expression (Table 3). One cell line, CMT47, was designated Luminal A subtype as the cells expressed low levels of HER2 and high levels of progesterone receptor but no estrogen receptor. Two CMT lines were designated Luminal B subtype (CMT12 and CMT119) as they expressed HER2 at high levels as well as lower or equal amounts of estrogen or progesterone receptors. When EGFr, HER3, and HER4 expression was also considered, three CMT cell lines (CMT9, CMT27, and CMT119) expressed high levels of HER3, while four CMT lines expressed HER4 (CMT9, CMT27, CMT47, and CMT119).
An overall assessment of the CMT phenotypes was created based on the extended receptor expression profiles including EGFr, c-erbB-3/HER3, and c-erbB-4/HER4 receptor mRNA levels (Table 4). Both CMT9 and CMT27 cells were designated as HER2+/HER3+ as they expressed no estrogen or progesterone receptors but expressed high levels of both HER2 and HER3 genes. Additionally, only the Luminal B CMT119 cells expressed high levels of HER4 along with high levels of EGFr, HER2, and HER3 and equal levels of progesterone receptor but no estrogen receptor expression compared to those in CMECs. CMT119 cells were designated Luminal B EGFr+/HER2+/HER3+/HER4+ due to the broad overexpression of all members of the erbB gene family. Thus, the extended analysis of these additional receptors allowed identification of at least two new potential canine breast cancer subtypes, the HER2+/HER3+ and Luminal B pan-HER+ tumor phenotypes.
Alignments of nucleotide sequences of all amplicons revealed highly conserved coding sequences that were comparable to orthologous, previously published canine and human mRNA sequences (Figs. 4 and 5). This analysis also authenticated all canine mRNA sequences amplified based on expression in the CMT cell lines and distinguished them from orthologous human sequences and other canine gene family members.
Clinicians require a clear definition of cancer subtype diagnosis upon which to base treatment and development of prognoses. Precise determination of breast cancer subtype allows more personalized and precise treatment regimens to be applied with a higher anticipated likelihood of success and is the basis for enhanced precision medicine [514]. Assays reported here promote the use of similar approaches in the treatment of canine breast cancers so that clinical diagnoses will be more precise and informative with respect to specific expression profiles of key receptors reported to be important in the development of breast neoplasia. These assays will also promote development of subtype-specific treatments suitable for canine mammary cancers.
In this investigation, well-characterized CMT cell lines [25], CMEC, and normal canine fibroblasts (NCF) were used to model normal and breast carcinoma gene expression profiles based on the expression of six well-characterized surface receptors reported to promote breast cancer in humans [4]. The approach in this study was based on assumptions that subtypes present in canine breast cancer would be similar to those in human breast cancer. We have reported that CMT28 cells overexpressed HER2/c-erbB-2 compared to that in other CMT cells and speculated that transformation in CMT28 cells was promoted by upregulated expression of the HER2/c-erbB-2 receptor pathway, making them relatively independent of growth factor stimulation [3]. Continuing characterization of genetic defects in these cell lines has focused on obtaining evidence of selection for atypical clones that are not representative of the primary tumor types. Although many of these cell lines have been in culture for multiple passages and no original primary tumor biopsy samples exist, comparison to fresh biopsy specimens from canine patients has confirmed the conserved nature of many of the primary genetic defects, especially those associated with the cyclin-dependent kinase tumor suppressor gene p16/INK4A [1819]. The data presented here demonstrate that this similarity can be extended beyond the tumor suppressor genes to include an array of cell surface receptors, including those reported to promote breast cancer in humans.
We have identified three of four key breast cancer subtypes among the CMT cell lines assessed including Luminal A, Luminal B, and HER2+ cell types. Additionally, we extended the analysis to include EGFr, HER3, and HER4 gene expression profiles thereby allowing identification of new subtypes of HER2+ cells expressing either EGFr or HER3 at levels higher than those in normal breast epithelial cells. This expanded analysis was included because, as noted, c-erbB-3 and c-erbB-4 expression has been implicated in breast cancer oncogenesis through heterodimer formation with the HER2 receptor [4]. We also identified a subtype of Luminal B that overexpresses all members of the erbB receptor gene family including EGFr, HER2, HER3, and HER4 as well as progesterone receptor. Expression of these molecules is likely key to understanding the mechanisms by which HER2 genes transform normal breast epithelium, as there is no known ligand for the HER2 receptor, and it is thought to activate through the formation of heterodimers with other members of the erbB family, particularly HER3 [4]. The current results demonstrate that these same mechanisms are also plausible neoplastic promoters in canine disease and may provide important new targets promoting development of improved diagnoses and new therapeutic strategies. Recent reports suggesting that HER2 expression data from CMT cells may not be reliable highlight the importance of careful validation of any QrtPCR assay by performing sequencing rather than by relying on antibodies of unknown specificity [10].
It was initially surprising that no triple-negative cell lines were discovered among those investigated, even though several lines, derived from highly metastatic tumors, were included. Recent reports have suggested that triple-negative breast cancers in dogs may be confined to inflammatory phenotypes [917]. Such tumors have not been part of our cell line development strategy and may explain the failure to recover this subtype in canine specimens. Future expansion of this strategy will be required to resolve this issue.
The spectrum of genes, reported to be defective in canine breast cancers, is expanding and becoming better defined, as has happened in human disease. Recent application of genome-wide association study (GWAS) analysis to canine patient populations has also clearly demonstrated the canine breed structure can promote this analysis, especially when coupled to robust comparative oncology [2629]. Identification of putative regulatory differences and changes to expression profiles that promote cancer risk have been inferred and these differences appear to be shared by otherwise unrelated canine breast cancers. Combining such analyses with known expression risk factors, such as those reported here, should promote more precise and personalized diagnosis of breast cancer in both species. Our previous discoveries of common tumor suppressor cyclin-dependent kinase inhibitor defects in canine mammary cancers, especially in the combined p16/INK4A and p14ARF locus, are important examples of approaches that can provide enhanced diagnostic power and point the way toward effective therapeutic targets [212181920].
Figures and Tables
 | Fig. 1Canine mammary epithelial cells (CMEC) and canine mammary tumor (CMT) cells. Phase contrast images of live CMT cells in culture were taken at 50–100% confluence in culture media by using a photomicroscope. The CMT primary cell image shows a representative colony of mammary carcinoma cells surrounded by a field of mammary fibroblasts prior to high-speed cell sorting on a MoFlo flow cytometer and cell sorter (Beckman Instruments). Magnifications for CMT12 and CMT27 cells were the same as CMT primary cell cultures. Representative images are shown. Scale bars = 1,000 µm (A and B), 400 µm (E and G), 200 µm (F and H). |
 | Fig. 2Expression of canine estrogen receptor α and progesterone receptor genes in canine mammary tumor (CMT) Cells. Quantitative reverse transcriptase polymerase chain reaction (QrtPCR) assays were developed to amplify discreetly each canine receptor mRNA in order to assess expression levels for each receptor in total cell RNA isolated from CMT cell lines CMT9, CMT12, CMT27, CMT28, CMT47, and CMT119. Agarose gel electrophoresis of semi-quantitative rtPCR for each amplicon is also shown to validate the QrtPCR values. (A) mRNA levels of estrogen receptor α (ER1) assessed by QrtPCR assay and normalized, for recovery and assay artifact, to expression levels of GAPDH and ribosomal protein L37 transcripts (L37). Normalized values were then expressed as a relative fold-function of the expression levels observed in normal canine mammary epithelial cells (CMEC), which were set to a value of 1 in the same assay. (B) mRNA levels of progesterone receptor (PR) were assessed by QrtPCR assay and calculated as described for ER1 mRNA expression. Agarose gels showing semi-quantitative levels of ERα and PR transcripts as well as GAPDH and L37 transcripts, which served as positive controls used for normalization, are also shown as positive controls to demonstrate the consistency of expression across samples. Significant differences, at the confidence levels (p values) noted, between CMEC and CMT expression levels are noted. An asterisk indicates significance at p≤0.05 or 0.01 as noted. FC, fold-change relative to control cells (CMEC); Neg, not expressed/detected in semi-quantitative or QrtPCR assays. |
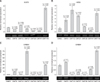 | Fig. 3Expression of canine c-erbB-1, c-erbB-2 (cHER2), c-erbB-3, and c-erbB-4 receptor genes in canine mammary tumor (CMT) cells. Quantitative reverse transcriptase polymerase chain reaction (QrtPCR) assays were developed to amplify discreetly each canine receptor gene to assess mRNA expression profiles of each receptor in total CMT cell RNA of cell lines CMT9, CMT12, CMT27, CMT28, CMT47, and CMT119. Agarose gel electrophoresis of semi-quantitative rtPCR for each amplicon is also shown to validate loading and as positive controls. The mRNA levels of (A) erbB-1/EGFr, (B) erbB-2/HER2, (C) erbB-3/HER3, and (D) erbB-4/HER4 are shown. Expression profiles were assayed by QrtPCR. Normalized levels calculated and significance determined as described for ER1 mRNA expression. |
 | Fig. 4ER/PR receptor family mRNA sequence analysis. mRNA sequences of ER1 and PR obtained from individual PCR products and derived amino acid sequences were compared. (A) Coding sequences (CDS) of ER1 from CMEC and canine mammary tumor (CMT)12 were aligned with published canine (GenBank No. XM_533454) and human (GenBank No. NM_000125) ER1 mRNA sequences. Inferred amino acid (AA) sequence in bold letters indicates differences between human and canine ER1 sequences. (B) Coding sequences of PR mRNAs from CMEC, CMT9, CMT27, CMT28, CMT47 and CMT119 were aligned with published canine (GenBank No. NM_001003074) and human (GenBank No. M15716) mRNA sequences and the inferred amino acid sequence is noted. |
 | Fig. 5c-erbB 1-4 receptor family mRNA sequence analysis. DNA sequencing of rtPCR amplicons of coding sequences of (A) c-erbB-1/EGFr, (B) c-erbB-2/HER2, (C) c-erbB-3/HER3, and (D) c-erbB-4/HER4 from canine mammary epithelial cells (CMEC) and canine mammary tumor (CMT) cell lines were aligned with corresponding canine and human sequences. Inferred amino acids (AA) in bold letters indicate differences between human and canine erbB sequences. GenBank accession Nos: canine c-erbB-1 No. XM_533073; human erbB-1 No. U48722; canine c-erbB-2 No. NM_001003217; human HER2 No. NM_004448; canine c-erbB-3 No. XM_538226; human erbB-3 No. NM_001982; canine c-erbB-4 No. XM_003640190; and human erbB-4 No. L07868. |
Table 3
Development of quantitative reverse transcriptase polymerase chain reaction human-like BrCa phenotypes for canine mammary tumor (CMT) cells

Table 4
Breast cancer phenotypes for canine mammary tumor (CMT) cells

HER3+ − erbB-3 expression levels ≥ canine mammary epithelial cells (CMEC). EGFr+/HER2+/HER3+/HER4+ − EGFr, erbB-2/HER2, erbB-3, and erbB-4 expression levels ≥ CMEC. *Basic human breast cancer phenotypes plus reference to overexpression of genes encoding EGFr/c-erbB-1 (2 CMT lines), c-erbB-3/HER3 (3 CMT lines), and c-erbB-4/HER4 (1 CMT line).
Acknowledgment
The authors thank Dr. B.F. Smith and Dr. A. Smith for valued discussions and critical reading of the manuscript and gratefully acknowledge the research support from the Auburn University Research Initiative in Cancer (AURIC) Program. Dr. F.M. Lutful Kabir was supported by an AURIC Hubbard Fellowship. Dr. F.M. Lutful Kabir and Dr. P. Agarwal were partially supported by Auburn University Cell and Molecular Biosciences.
References
1. Ades F, Zardavas D, Bozovic-Spasojevic I, Pugliano L, Fumagalli D, de Azambuja E, Viale G, Sotiriou C, Piccart M. Luminal B breast cancer: molecular characterization, clinical management, and future perspectives. J Clin Oncol. 2014; 32:2794–2803.

2. Agarwal P, Sandey M, DeInnocentes P, Bird RC. Tumor suppressor gene p16/INK4A/CDKN2A-dependent regulation into and out of the cell cycle in a spontaneous canine model of breast cancer. J Cell Biochem. 2013; 114:1355–1363.

3. Ahern TE, Bird RC, Bird AEC, Wolfe LG. Expression of the oncogene c-erbB-2 in canine mammary cancers and tumor-derived cell lines. Am J Vet Res. 1996; 57:693–696.
4. Arteaga CL, Engelman JA. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014; 25:282–303.

5. Beck J, Hennecke S, Bornemann-Kolatzki K, Urnovitz HB, Neumann S, Ströbel P, Kaup FJ, Brenig B, Schütz E. Genome aberrations in canine mammary carcinomas and their detection in cell-free plasma DNA. PLoS One. 2013; 8:e75485.

6. Bird RC, Bird AEC, DeInnocentes P. Animal cell separation and subcellular fractionation. Encyclopedia of Life Sciences (ELS). Chichester: John Wiley & Sons;2011.
7. Bird RC, DeInnocentes P, Bird AEC, van Ginkel FW, Lindquist J, Smith BF. An autologous dendritic cell canine mammary tumor hybrid-cell fusion vaccine. Cancer Immunol Immunother. 2011; 60:87–97.

8. Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol. 2005; 23:7350–7360.

9. Burrai GP, Tanca A, De Miglio MR, Abbondio M, Pisanu S, Polinas M, Pirino S, Mohammed SI, Uzzau S, Addis MF, Antuofermo E. Investigation of HER2 expression in canine mammary tumors by antibody-based, transcriptomic and mass spectrometry analysis: is the dog a suitable animal model for human breast cancer? Tumour Biol. 2015; 36:9083–9091.

10. Caceres S, Peña L, de Andres PJ, Illera MJ, Lopez MS, Woodward WA, Reuben JM, Illera JC. Establishment and characterization of a new cell line of canine inflammatory mammary cancer: IPC-366. PLoS One. 2015; 10:e0122277.

11. Cullen JM, Page R, Misdorp W. An overview of cancer pathogenesis, diagnosis and management. In : Meuten DJ, editor. Tumors in Domestic Animals. 4th ed. Ames: Iowa State University Press;2002. p. 3–45.
12. DeInnocentes P, Agarwal P, Bird RC. Phenotype-rescue of cyclin-dependent kinase inhibitor p16/INK4A defects in a spontaneous canine cell model of breast cancer. J Cell Biochem. 2009; 106:491–505.

13. DeInnocentes P, Li LX, Sanchez RL, Bird RC. Expression and sequence of canine SIRT2 and p53 genes in canine mammary tumour cells - effects on downstream targets Wip1 and p21/Cip1. Vet Comp Oncol. 2006; 4:161–177.

14. Gama A, Alves A, Schmitt F. Identification of molecular phenotypes in canine mammary carcinomas with clinical implications: application of the human classification. Virchows Arch. 2008; 453:123–132.

15. Goldschmidt M, Peña L, Rasotto R, Zappulli V. Classification and grading of canine mammary tumors. Vet Pathol. 2011; 48:117–131.

16. Im KS, Kim NH, Lim HY, Kim HW, Shin JI, Sur JH. Analysis of a new histological and molecular-based classification of canine mammary neoplasia. Vet Pathol. 2014; 51:549–559.

17. Kim NH, Lim HY, Im KS, Kim JH, Sur JH. Identification of triple-negative and basal-like canine mammary carcinomas using four basal markers. J Comp Pathol. 2013; 148:298–306.

18. Lutful Kabir FM, Agarwal P, DeInnocentes P, Zaman J, Bird AC, Bird RC. Novel frameshift mutation in the p16/INK4A tumor suppressor gene in canine breast cancer alters expression from the p16/INK4A/p14ARF locus. J Cell Biochem. 2013; 114:56–66.

19. Lutful Kabir FM, Alvarez CE, Bird RC. Canine mammary carcinomas: a comparative analysis of altered gene expression. Vet Sci. 2016; 3:1.

20. Lutful Kabir FM, DeInnocentes P, Bird RC. Altered microRNA expression profiles and regulation of INK4A/CDKN2A tumor suppressor genes in canine breast cancer models. J Cell Biochem. 2015; 116:2956–2969.

21. Onitilo AA, Engel JM, Greenlee RT. Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin Med Res. 2009; 7:4–13.

22. Peña L, Gama A, Goldschmidt MH, Abadie J, Benazzi C, Castagnaro M, Díez L, Gärtner F, Hellmén E, Kiupel M, Millán Y, Miller MA, Nguyen F, Poli A, Sarli G, Zappulli V, de las Mulas JM. Canine mammary tumors: a review and consensus of standard guidelines on epithelial and myoepithelial phenotype markers, HER2, and hormone receptor assessment using immunohistochemistry. Vet Pathol. 2014; 51:127–145.
23. Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge Ø, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000; 406:747–752.

24. Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010; 12:R68.

25. Sassi F, Benazzi C, Castellani G, Sarli G. Molecular-based tumour subtypes of canine mammary carcinomas assessed by immunohistochemistry. BMC Vet Res. 2010; 6:5.

26. Schiffman JD, Breen M. Comparative oncology: what dogs and other species can teach us about humans with cancer. Philos Trans R Soc Lond B Biol Sci. 2015; 370:20140231.

27. Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE, Børresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001; 98:10869–10874.

28. Su S, Bird RC. Cell cycle, differentiation and tissue-independent expression of ribosomal protein L37. Eur J Biochem. 1995; 232:789–797.

29. Tonomura N, Elvers I, Thomas R, Megquier K, Turner-Maier J, Howald C, Sarver AL, Swofford R, Frantz AM, Ito D, Mauceli E, Arendt M, Noh HJ, Koltookian M, Biagi T, Fryc S, Williams C, Avery AC, Kim JH, Barber L, Burgess K, Lander ES, Karlsson EK, Azuma C, Modiano JF, Breen M, Lindblad-Toh K. Genome-wide association study identifies shared risk loci common to two malignancies in golden retrievers. PLoS Genet. 2015; 11:e1004922.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download