Abstract
Recent developments in genome editing technology using meganucleases demonstrate an efficient method of producing gene edited pigs. In this study, we examined the effectiveness of the transcription activator-like effector nuclease (TALEN) system in generating specific mutations on the pig genome. Specific TALEN was designed to induce a double-strand break on exon 9 of the porcine α1,3-galactosyltransferase (GGTA1) gene as it is the main cause of hyperacute rejection after xenotransplantation. Human decay-accelerating factor (hDAF) gene, which can produce a complement inhibitor to protect cells from complement attack after xenotransplantation, was also integrated into the genome simultaneously. Plasmids coding for the TALEN pair and hDAF gene were transfected into porcine cells by electroporation to disrupt the porcine GGTA1 gene and express hDAF. The transfected cells were then sorted using a biotin-labeled IB4 lectin attached to magnetic beads to obtain GGTA1 deficient cells. As a result, we established GGTA1 knockout (KO) cell lines with biallelic modification (35.0%) and GGTA1 KO cell lines expressing hDAF (13.0%). When these cells were used for somatic cell nuclear transfer, we successfully obtained live GGTA1 KO pigs expressing hDAF. Our results demonstrate that TALEN-mediated genome editing is efficient and can be successfully used to generate gene edited pigs.
Due to anatomical and physiological similarities with humans and high production efficiency, pigs can be used as model animals for biomedical research [20]. In addition, xenotransplantation using pig organs is expected to become a suitable alternative that helps alleviate the shortage of human transplantation donors around the world. However, in pig-to-human xenotransplantation, the grafted organ is often rejected. Such rejection occurs via immune mechanisms in four stages, hyperacute rejection (HAR), acute xenograft rejection, cell mediated rejection and chronic rejection [29]. First, grafts from pigs can be immediately rejected by the human immune system in xenotransplantation via HAR. This type of rejection is mainly caused by the xenoantigen of galactose-α1,3-galactose (αGal), which leads to severe graft destruction characterized by thrombosis, edema and cell infiltration within minutes to hours, and is known to be a major hurdle to pig-to-primate xenotransplantation. Thus, disruption of the α1,3-galactosyltransferase (GGTA1) gene, which is essential for αGal synthesis, is the first step toward overcoming HAR. There have been several attempts to generate GGTA1 knockout (KO) swine by several groups through integration of traditional DNA homologous recombination (HR) and somatic cell nuclear transfer (SCNT) [121420]. Using these GGTA1 KO pigs, subsequent studies found that pig-to-primate translation of hearts can prolong the survivability period of the xenograft [13]. Moreover, the complement system in the human body distinguishes foreign objects from the self via a range of specialized cell-surface and soluble proteins. These regulatory proteins include human decay-accelerating factor (hDAF), membrane cofactor protein (MCP), and cluster of differentiation 59 (CD59). Therefore, the development of αGal deficient pigs expressing human complement regulatory proteins such as hDAF, MCP and CD59 is necessary to overcome HAR caused by transplantation of pig organs into humans.
Since production of the first transgenic cloned pigs with a targeted modification, various transgenic pigs have been generated. However, the production of desired gene targeted pigs has been extremely rare until recently because of the lack of effective technologies. Recently, application of artificially engineered nucleases such as zinc-finger nuclease (ZFN), transcription activator-like effector nuclease (TALEN), and the clustered regularly-interspaced short palindromic repeats/CRISPR associated protein 9 (CRISPR/Cas9) system in transgenic pig production has enabled generation of KO animals at high efficiency [8]. Sequence-specific artificially engineered nucleases such as ZFN, TALEN and the CRISPR/Cas9 system can cleave DNA in mammalian cells, and, as a consequence, induce HR more effectively than conventional methods [9]. In contrast to an extremely low gene targeting efficiency from the traditional gene targeting approach through HR methods, use of ZFN has been shown to be a more efficient method for production of gene KO pigs [10]. However, ZFN requires more optimization in terms of design and the assembly for successful gene targeting, and may also be restricted to some genomic target sites. When compared to ZFN, TALEN as a next generation genome-modifying technology has high scientific visibility for genetic engineering in various animals and can cause DNA double-strand breaks in a specific desired sequence, resulting in mutation of target genes at higher efficiency than ZFN. Moreover, TALEN has some advantages such as availability [4], specificity [3], flexibility and lower toxicity [18] than ZFN. To date, TALEN has been successfully applied for efficient gene targeting in several animal models [2611182422], including porcine KO models [41719252830].
Establishing cell lines with targeted disruption through suitable selection of cell colonies is a very important process. Generally, the antibiotic selection approach has been used to screen targeted cells; however, the integration of antibiotic genes is unavoidable and may therefore cause concerns regarding the food safety of the animals and unexpected biological effects. Accordingly, gene targeting without exogenous DNA integration is worth pursuing. Recently, the use of magnetic beads was suggested as an improved strategy for selection of αGal-KO cells [21]. The efficiency of the number of GGTA1 null colonies was increased from 0.01% to 4.5% in that study. A similar technique was used in this study that allows efficient selection of αGal-KO cells within a mixed population of cells by using magnetic beads.
In the present study, we attempted to disrupt the GGTA1 gene and express additional transgenes in pig somatic cells through a combination of TALEN-mediated gene modification and SCNT technology. We have successfully produced pigs that lack GGTA1 and express hDAF. Phenotypic and functional analyses of the mutated pigs showed that production of GGTA1 KO pigs expressing human complement regulatory proteins such as hDAF provides a more advanced organ source for xenotransplantation research.
All pigs were raised at the MGENPLUS Biotechnology Research Institute, Seoul, Korea. All animal experiments were approved by the Institutional Animal Care and Use Committee of the institute of MGENPLUS Biotechnology Research Institute, and all procedures followed the guidelines of the Committee.
TALEN expression constructs were assembled by the ToolGen company (Korea). A pair of TALEN constructs was designed to target a region on exon 9 of the porcine GGTA1 genes (ensemble ID: ENSSSCG00000003385). The GGTA1 TALEN binding sequences were as follows: left pair 5'-TTCTTAATATCTGCAAATAC-3' and right pair 5'-GGCCACAAAGTCATCTTTTA-3'. There is a 13-bp spacer between the left and right TALEN. The TALEN constructs were transfected into porcine fetal fibroblasts (PFFs) to target GGTA1. hDAF expression vector was constructed as follows to produce human DAF: expression of the hDAF gene is derived by CMV promoter. An insert (2.2 kb) consisting of CMV promoter-hDAF-polyA cassette including the AvrII-XmaI fragment from hDAF mRNA (accession No. NM_000574.4) was ligated into the pCR2.1-TOPO plasmid (Invitrogen, USA).
PFFs isolated from 35-day-old PWG miniature pig fetuses (male and female) were used in this study. Two days before transfection, PFFs were cultivated in tissue culture dish. When the cells reached 80% confluency, TALEN was transfected into these cells by electroporation using Amaxa 3D-Necleofector (Lonza, Switzerland) and an hDAF expression vector was also co-transfected to express hDAF protein. Two or three days after transfection, cells were subjected to MACS sorting based on the biotin-labeled IB4 lectin attached to dynabeads magnetic beads [10]. Selected cells were plated in 6 well plates and cultured as a form of single cell in cell culture medium. Next, cell colonies derived from single cells were separated mechanically and a part of the separated colony was genotyped by PCR and subsequent sequencing. Cell colonies with the desired mutations were cultured and used as donor cells for SCNT.
αGal-negative cells were selected by magnetic beads (Dynabeads; Invitrogen, USA) as previously described [10]. Selection was performed on cells using biotin-conjugated IB4 lectin attached to streptavidin-coated magnetic beads. Cells were harvested and suspended at 5 × 106 cells/mL in phosphate-buffered saline containing 5 µg biotin-conjugated IB4 lectin for 15 min on ice, then incubated with 0.5 mg Dynabeads M-280 streptavidin (Invitrogen) for 30 min. αGal-positive cells were removed using a magnetic rack for 2 min. Gal-negative cells were collected from the supernatant, plated and incubated until colonies developed.
The genomic DNA of each cell colony was extracted using a DNA extraction kit (Qiagen, Germany) according to the manufacturer's instructions. PCR was used for genotyping. To detect mutations on GGTA1, the following primers were used: left primer 5'-GCTGTTACAGCAACAGACGTCTC-3' and right primer 5'-GCATGCAGAAGAGGAAGTCCACC-3'. Mutations on the GGTA1 gene were assessed by a T7 endonuclease I (T7E1) assay. Briefly, the purified PCR products from the DNA isolated from colonies were denatured at 95℃ for 5 min, then re-annealed at room temperature for 10 min, after which they were digested by T7E1 (ToolGen, Korea) at 37℃ for 30 min. Digestion of the PCR product was expected if the colony contained mutated GGTA1. Conditions for PCR amplification were one cycle of initial-denaturation at 94℃ for 5 min followed by 30 cycles of denaturation at 95℃ for 60 sec, annealing at 55℃ for 60 sec, and elongation at 72℃ for 60 sec, and then one cycle of post-elongation at 72℃ for 10 min. PCR products with potential modifications on GGTA1 were confirmed by sequencing. For detection of DAF insertion on the genomic region (926 bp) and mRNA expression (157 bp), the following primers were used: forward primer 5'-AATTCCTGGCGAGAAGGACT-3' and reverse primer 5'-GCAAGCCCATGGTTACTAGC-3' (on genomic region), forward primer 5'-CAGCACCACCACAAATTGAC-3' and reverse primer 5'-CCACTCCACTCTCCTTCATCA-3' (for mRNA expression). Conditions for PCR amplification were one cycle of initial-denaturation at 94℃ for 5 min followed by 35 cycles of denaturation at 94℃ for 30 sec, annealing at 60℃ for 30 sec, and elongation at 72℃ for 60 sec, and one cycle of post-elongation at 72℃ for 5 min.
Pig ovaries were collected from a local abattoir and transported to the laboratory in 0.9% (w/v) NaCl solution at 25 to 30℃. Follicular contents from antral follicles (3–6 mm in diameter) were aspirated using an 18-gauge needle attached to a 10 mL disposable syringe. The contents were pooled in a conical tube at 39℃ and allowed to settle for a few minutes. The sediment was then aspirated and diluted with TL-Hepes containing 100 U/mL penicillin G and 100 mg/mL streptomycin sulphate (pen-strep; Invitrogen). Cumulus-oocyte complexes (COCs) with intact compact cumulus cell layers were selected and washed three times in TL-Hepes before being transferred to a modified TCM-199 (Invitrogen) supplemented with 10 ng/mL EGF, 26 mM NaHCO3, 3.05 mM glucose, 0.57 mM cystine, 0.91 mM sodium pyruvate, 1% (v/v) pen-strep, 0.5 µg/mL follicle stimulating hormone, 0.5 µg/mL luteinizing hormone and 10% porcine follicular fluid. After 22 h of maturation, COCs were transferred to a maturation medium without gonadotropins. The COCs were cultured at 39℃ under 5% CO2 at 100% humidity. After 44 h of maturation, cumulus cells surrounding the oocytes were removed by pipetting with 0.1% hyaluronidase in TL-Hepes supplemented with 0.1% polyvinyl alcohol. Denuded oocytes with polar bodies were then selected and used for SCNT.
Enucleation and donor cell injection procedures in SCNT were conducted on an inverted microscope (Nikon, Japan) equipped with a micromanipulator (Nikon-Narishige, Japan). Matured oocytes were enucleated by aspirating the first polar body and adjacent cytoplasm with a beveled glass pipette (20 µm internal diameter) in manipulation medium supplemented with cytochalasin B (5 mg/mL stock, 1.5 µL per 10 mL manipulation medium). A single donor cell was injected into the perivitelline space of enucleated oocytes. Oocyte cytoplasm-cell complexes were then fused and activated by electric pulse (two DC pulses of 1.1 kV/cm for 60 µsec; ECM 2001; BTX/Harvard Apparatus, USA) in fusion medium with 0.3 M mannitol and 0.1 mM CaCl2. Reconstructed embryos were cultured in porcine zygote medium under 5% CO2 at 39℃ until embryo transfer. About 200 embryos were surgically transferred into the oviduct of a surrogate at day two after estrus was observed. Early pregnancy was detected by ultrasound at day 28 after embryo transfer. After an approximately 114 day gestation period, natural delivery was conducted. On the day of birth, a tail biopsy was performed from each piglet for genomic DNA extraction and genotyping.
A complement lysis assay of skin fibroblasts of six cloned pigs and monoallelic or biallelic GGTA1 KO cells derived from this study and wild-type cells (control) was conducted. The experiment was performed as previously described [28]. Briefly, cells at 70 to 80% confluency in 24-well plates were incubated with 250 µL of 2 µg/mL calcein AM (Molecular Probes, USA) for 1 h at 38℃ under 5% CO2, then washed with PBS. Cells were mixed with 250 µL of 20% normal human serum (NHS; EMD Millipore, USA) and 0.5% bovine serum albumin (BSA) diluted in dextrose-gelatin-veronal solution (Lonza). Next, cells were incubated for 1 h at 38oC and 5% CO2. Cell supernatants were collected into a 96-well plate (Sample 1), and 250 µL of 0.1% Triton X-100 was mixed into each well and incubated for 15 min at room temperature to lyse the remaining cells. Next, supernatants containing lysed cells were transferred to a 96-well plate (Sample 2), after which the quantity of calcein release was determined by fluorescein detection using a microplate reader (VICTOR3; PerkinElmer, USA) at 480 nm/535 nm (excitation/emission wavelength). The ratio of cytotoxicity was calculated using the following formula: Cytotoxicity (%) = (Sample 1)/(Sample 1 + Sample 2) × 100%. Cytotoxicity at NHS was normalized to cytotoxicity at BSA.
A TALEN pair targeting exon 9 of the GGTA1 gene was introduced to PFFs by electroporation. The construction of TALENs used in this study is shown in Fig. 1A. Application of the GGTA1 TALEN set yielded the formation of 53.7% (66/123) GGTA1 mutant colonies after selecting cells with a biotin-labeled IB4 lectin attached to magnetic beads. Among these mutant colonies, 35.0% (43/123) carried a biallelic modification of GGTA1, which was confirmed by DNA sequencing of the TALEN binding sites (Table 1). Moreover, a TALEN pair targeting GGTA1 was co-transfected with the hDAF expression vector into PFFs by electroporation. Two days after transfection, single cells were sorted using a biotin-labeled IB4 lectin attached to magnetic beads. Single cell-derived colonies were selected and screened by the T7E1 assay and PCR DNA sequencing (panel B and C in Fig. 1). A total of 46 colonies were obtained from these, of which, six were shown to be mutated by the T7E1 assay (panel B in Fig. 1). Sequencing revealed that the colonies carried mono-allelic mutation of GGTA1 and random insertion of hDAF DNA. One allele of all colonies harbored some deletion (panel C in Fig. 1). The results showed that the TALEN-mediated targeting efficiency of co-transfection with hDAF expression vector is 13% (6/46). PCR amplification and subsequent DNA sequencing, which was used to determine mutation at the target site, showed various biallelic mutation patterns, including small deletions (e.g., −1 base), large deletions (e.g., −150 base), small insertions (e.g., +4 base) and base substitutions (e.g., 1 base), which are characteristic of non-homologous end joining-induced DNA damage responses (data not shown).
Female PFFs expressing hDAF were used as donor cells for SCNT and embryo transfer to generate GGTA1 KO pigs. A total of 3,381 embryos were generated and transferred to 12 surrogates (average 280 embryos/surrogate), and three pregnancies were established. Six live-born piglets and three stillborn piglets were obtained from these pregnancies. Genotyping of the piglets showed eight base mono-allelic mutations of GGTA1 and hDAF expression (Fig. 2). Sequencing of the genome revealed a heterozygous mutation with an eight base deletion in one allele of the target loci. Unfortunately, the one mono-allelic mutant live-born piglet died within two days of birth. No obvious defects of organs were observed in the dead piglet.
Organ tissues were dissected from the deceased cloned piglets and skin tissue was collected from the ear and tail of all live cloned piglets. To examine the genotype of the cloned fetuses and piglets, a region of GGTA1 flanking the TALEN target region was amplified by PCR, subcloned into a vector, and sequenced. Mutations were found at the TALEN target region in cloned fetuses and piglets (Fig. 2). Genomic PCR analysis and RT-PCR analysis were conducted to confirm insertion and expression of hDAF in all cloned piglets. The results showed that hDAF was inserted into a genomic region of cloned pigs and expressed in some tissues, suggesting the successful generation of αGal KO piglets expressing hDAF protein (Fig. 2).
The resistance for immune reaction by the complement system in human serum was tested in transgenic cells and skin fibroblasts from cloned piglets. These cells (No. 1–6) have one mutant allele for GGTA1 genes and also expressed the hDAF gene. As shown in Fig. 3, transgenic cells were more resistant to lysis when exposed to human serum than control cells. The ratio of cytotoxicity of cells treated with normal human serum was 0.90 ± 0.01 (No. 1), 1.52 ± 0.08 (No. 2), 1.47 ± 0.01 (No. 3), 1.44 ± 0.05 (No. 4), 1.37 ± 0.04 (No. 5) and 1.50 ± 0.02 (No. 6) when compared with cells treated with BSA (1.00). However, cytotoxicity of wild type control and GGTA1 KO cells in the normal human serum were 1.68 ± 0.02, 1.52 ± 0.02 (monoallelic KO) and 1.48 ± 0.01 (biallelic KO), respectively (Fig. 3).
In this study, we successfully disrupted the GGTA1 gene in pig somatic cells. Genotyping of the GGTA1 KO cells indicates a higher frequency of deletion mutants than insertion mutants. This is consistent with the results of other studies in which TALEN was utilized to induce targeted modification of the genome in embryos [24].
Because of physiological and anatomical similarities with humans, pigs have been considered to be more appropriate experimental models than small animal models such as rodents for studying human diseases and are also potential xenogenic donors for organ-transplantation. However, difficulty in inducing specific genetic modification has prevented the application of pigs in biomedical research and drug discovery. In the current study, we successfully generated GGTA1 KO pigs at high efficiency using TALEN technology combined with SCNT. Our approaches established in this study increased the efficiency of gene targeting in primary pig cells by more than ten times when compared with the traditional HR method for gene targeting. In addition, we have provided a high-efficiency method for generating gene-targeted transgenic pigs for various purposes.
The use of TALEN technology can support the generation of KO models in other large animals in which there are no true ES cell lines available. Injection of TALEN into embryos generated targeted mutations in zebrafish [11], Xenopus [18], rats [26], mice [24] and rabbits [22]. There are also reports of KO swine produced with TALENS [41719252830]. However, animals produced from TALEN injected embryos have chimeric mutations with multiple sites [22]. Because this approach requires more breeding rounds to obtain an animal with a single mutation, it is not desirable. A recent report suggested that injection of embryos with artificially engineered nucleases can be effective resulting in lower mosaic [27], but the approach is not publically available. Therefore, we used somatic cells in gene targeting with the TALEN technology. The generated somatic cells were subsequently used in SCNT to produce GGTA1 KO pigs. An hDAF expression vector was co-transfected into primary fetal fibroblasts during the gene targeting process to integrate hDAF. The gene targeting efficiency of TALEN pairs in somatic cells was 53.7%, which was significantly higher efficiency than that of traditional methods utilizing endogenous homologous recombination. Moreover, we obtained many biallelic KO cells in the GGTA1 locus, which was nearly impossible using traditional methods employing DNA HR, and the generation rate was up to 35.0%. TALEN-mediated genome engineering clearly has the potential to revolutionize genetics and genome engineering in pigs by introducing a variety of genomic changes, including monoallelic KO, biallelic KO, large chromosomal deletions/inversions and potentially, precise allelic introgression. TALEN can also be easily designed and assembled using molecular biology techniques available in most laboratories. Accordingly, TALEN is a reliable gene targeting tool capable of inducing highly efficient genetic modifications in porcine somatic cells.
In the present study, we attempted to isolate GGTA1 KO cells from transfected cell populations using magnetic beads as previously described [10]. The method for selecting GGTA1 KO cells developed in this study overcame problems of time and complexity encountered in previous methods, which commonly use antibiotics selection. The use of antibiotics requires an extended period of culture period in vitro. Because of these high passage numbers and longer culture period, donor cell conditions worsen, resulting in lower embryo development. However, this IB4-lectin-mediated method for selection of αGal epitope-negative cells provides an advantage to the previous method. Because selection methods used in this study do not require antibiotic selection of modified cells, this method provides higher efficiency and requires less time than the previous selection method. Overall, we efficiently selected αGal epitope-negative cells with heterozygous and homozygous KO of the GGTA1 gene by removing αGal epitope-expressing cells from transfected cell populations.
GGTA1, which can be responsible for HAR, is expressed on endothelial cells and produced the α1, 3Gal carbohydrate on the cell surface [671629]. Thus, GGTA1 KO pigs have been used consistently for pig-to-primate xenotransplantation. In some studies, low levels of αGal antigen in the GGTA1 KO pig cells reduced HAR somewhat in the pig-to-primate xenotransplantation. Moreover, expression of hDAF genes in pig cells for regulation of the human complement system on the cell surface provided effective protection against HAR in xenotransplantation [5]. Expression of human complement regulatory proteins, such as hDAF, MCP and CD59, on endothelial cells, was critical for protection against HAR in xenotransplantation because the endothelial cells are first exposed to the various components of the recipient's immune system [1]. Peripheral blood mononuclear cells from hDAF transgenic pigs had higher survival rates when treated with complement in human serum [15]. Transgenic pigs expressing the hDAF gene were effectively protected against HAR in xenotrasplantation by regulation of the complement system on the cell surface [2523]. In this study, expression of hDAF protected against human normal serum more than αGal KO, as in previous studies. Therefore, development of GGTA1 KO pigs that express hDAF genes is important in pig-to-primate xenotrasplantation. However, according to the multiple expression condition of hDAF in tissue from cloned piglets by random integration into the genome, the cytotoxicity of samples differed. So, further study should be demanded as stably elevated expression of hDAF.
We successfully produced live piglets from GGTA1 KO and hDAF expressing cells through SCNT. Genotype assay confirmed that the mutation pattern in the cloned piglets was consistent with that in the donor cells. Disruption of the GGTA1 gene and expression of hDAF in cloned pigs led to their regulation of the function of the complement system through complement-mediated lysis assay with human complement serum. Cells obtained from the GGTA1 (-/+) and hDAF expressing piglets were more resistant to lysis by pooled complement-preserved NHS than those from wild-type pigs, which suggests that the organs from these pigs can overcome HAR upon transplantation into humans and primates. In conclusion, we demonstrated that TALEN editing technology can be successfully applied to pigs to produce live gene edited pigs. The results of our study also indicate that TALENs can be used efficiently for precise gene modification in other large animals. GGTA1 mutated piglets expressing hDAF will provide a new organ source for xenotransplantation research.
Figures and Tables
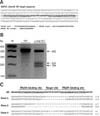 | Fig. 1Generation of porcine α1,3-galactosyltransferase (GGTA1) knockout (KO) fibroblasts with transcription activator-like effector nucleases (TALENs). (A) Sequences of the TALEN binding site in the GGTA1 gene. (B) TALEN driven GGTA1 mutations detected by the T7 endonuclease I (T7E1) assay in a cell population isolated using a biotin-labeled IB4 lectin attached to dynabeads magnetic beads. (C) DNA sequencing of TALEN target region in transfected cells. WT, wild type. |
 | Fig. 2Production of TALEN-mediated piglets by somatic cell cloning. (A) DNA sequencing analysis of GGTA1 TALEN target region in wild-type cells, cloned fetuses and cloned piglets. (B) Genomic PCR of hDAF in cloned piglets. (C) RT-PCR of hDAF in cloned piglets. (D) GGTA1 KO pig expressing hDAF generated from TALEN-mediated donor cells. |
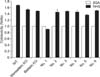 | Fig. 3Complement-mediated lysis assay. Monoallelic KO indicates GGTA1 KO heterozygous cells and biallelic KO indicates GGTA1 KO homozygous cells. From number 1 to 6 are cloned piglets derived from hDAF expressing GGTA1 KO cells by nuclear transfer. BSA, bovine serum albumin; NHS, normal human serum. |
Acknowledgments
This paper was partially supported by Sunchon National University Research Fund in 2013.
References
1. Aigner B, Klymiuk N, Wolf E. Transgenic pigs for xenotransplantation: selection of promoter sequences for reliable transgene expression. Curr Opin Organ Transplant. 2010; 15:201–206.

2. Baldan N, Rigotti P, Calabrese F, Cadrobbi R, Dedja A, Iacopetti I, Boldrin M, Seveso M, Dall'Olmo L, Frison L, De Benedictis G, Bernardini D, Thiene G, Cozzi E, Ancona E. Ureteral stenosis in HDAF pig-to-primate renal xenotransplantation: a phenomenon related to immunological events? Am J Transplant. 2004; 4:475–481.

3. Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010; 48:419–436.
4. Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci U S A. 2012; 109:17382–17387.

5. Chen G, Sun H, Yang H, Kubelik D, Garcia B, Luo Y, Xiang Y, Qian A, Copeman L, Liu W, Cardella CJ, Wang W, Xiong Y, Wall W, White DJ, Zhong R. The role of anti-non-Gal antibodies in the development of acute humoral xenograft rejection of hDAF transgenic porcine kidneys in baboons receiving anti-Gal antibody neutralization therapy. Transplantation. 2006; 81:273–283.

6. Galili U. Interaction of the natural anti-Gal antibody with alpha-galactosyl epitopes: a major obstacle for xenotransplantation in humans. Immunol Today. 1993; 14:480–482.

7. Galili U, Shohet SB, Kobrin E, Stults CLM, Macher BA. Man, apes, and Old World monkeys differ from other mammals in the expression of α-galactosyl epitopes on nucleated cells. J Biol Chem. 1988; 263:17755–17762.

8. Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Ménoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009; 325:433.

9. Gouble A, Smith J, Bruneau S, Perez C, Guyot V, Cabaniols JP, Leduc S, Fiette L, Avé P, Micheau B, Duchateau P, Pâques F. Efficient in toto targeted recombination in mouse liver by meganuclease-induced double-strand break. J Gene Med. 2006; 8:616–622.

10. Hauschild J, Petersen B, Santiago Y, Queisser AL, Carnwath JW, Lucas-Hahn A, Zhang L, Meng X, Gregory PD, Schwinzer R, Cost GJ, Niemann H. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc Natl Acad Sci U S A. 2011; 108:12013–12017.

11. Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011; 29:699–700.

12. Kolber-Simonds D, Lai L, Watt SR, Denaro M, Arn S, Augenstein ML, Betthauser J, Carter DB, Greenstein JL, Hao Y, Im GS, Liu Z, Mell GD, Murphy CN, Park KW, Rieke A, Ryan DJJ, Sachs DH, Forsberg EJ, Prather RS, Hawley RJ. Production of α-1,3-galactosyltransferase null pigs by means of nuclear transfer with fibroblasts bearing loss of heterozygosity mutations. Proc Natl Acad Sci U S A. 2004; 101:7335–7340.

13. Kuwaki K, Tseng YL, Dor FJMF, Shimizu A, Houser SL, Sanderson TM, Lancos CJ, Prabharasuth DD, Cheng J, Moran K, Hisashi Y, Mueller N, Yamada K, Greenstein JL, Hawley RJ, Patience C, Awwad M, Fishman JA, Robson SC, Schuurman HJ, Sachs DH, Cooper DKC. Heart transplantation in baboons using α1,3-galactosyltransferase gene-knockout pigs as donors: initial experience. Nat Med. 2005; 11:29–31.

14. Lai L, Kolber-Simonds D, Park KW, Cheong HT, Greenstein JL, Im GS, Samuel M, Bonk A, Rieke A, Day BN, Murphy CN, Carter DB, Hawley RJ, Prather RS. Production of α-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science. 2002; 295:1089–1092.

15. Lavitrano M, Bacci ML, Forni M, Lazzereschi D, Di Stefano C, Fioretti D, Giancotti P, Marfé G, Pucci L, Renzi L, Wang H, Stoppacciaro A, Stassi G, Sargiacomo M, Sinibaldi P, Turchi V, Giovannoni R, Della Casa G, Seren E, Rossi G. Efficient production by sperm-mediated gene transfer of human decay accelerating factor (hDAF) transgenic pigs for xenotransplantation. Proc Natl Acad Sci U S A. 2002; 99:14230–14235.

16. Le Bas-Bernardet S, Anegon I, Blancho G. Progress and prospects: genetic engineering in xenotransplantation. Gene Ther. 2008; 15:1247–1256.

17. Lee K, Kwon DN, Ezashi T, Choi YJ, Park C, Ericsson AC, Brown AN, Samuel MS, Park KW, Walters EM, Kim DY, Kim JH, Franklin CL, Murphy CN, Roberts RM, Prather RS, Kim JH. Engraftment of human iPS cells and allogeneic porcine cells into pigs with inactivated RAG2 and accompanying severe combined immunodeficiency. Proc Natl Acad Sci U S A. 2014; 111:7260–7265.

18. Lei Y, Guo X, Liu Y, Cao Y, Deng Y, Chen X, Cheng CHK, Dawid IB, Chen Y, Zhao H. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs). Proc Natl Acad Sci U S A. 2012; 109:17484–17489.

19. Lillico SG, Proudfoot C, Carlson DF, Stverakova D, Neil C, Blain C, King TJ, Ritchie WA, Tan W, Mileham AJ, McLaren DG, Fahrenkrug SC, Whitelaw CBA. Live pigs produced from genome edited zygotes. Sci Rep. 2013; 3:2847.

20. Phelps CJ, Koike C, Vaught TD, Boone J, Wells KD, Chen SH, Ball S, Specht SM, Polejaeva IA, Monahan JA, Jobst PM, Sharma SB, Lamborn AE, Garst AS, Moore M, Demetris AJ, Rudert WA, Bottino R, Bertera S, Trucco M, Starzl TE, Dai Y, Ayares DL. Production of α1,3-galactosyltransferase-deficient pigs. Science. 2003; 299:411–414.

21. Reyes LM, Estrada JL, Ivary B, Sidner RA, Paris LL, Tector AJ. Efficient selection of Gal-knockout pig cells for somatic cell nuclear transfer. J Surg Res. 2013; 184:e37–e42.

22. Song J, Zhong J, Guo X, Chen Y, Zou Q, Huang J, Li X, Zhang Q, Jiang Z, Tang C, Yang H, Liu T, Li P, Pei D, Lai L. Generation of RAG 1- and 2-deficient rabbits by embryo microinjection of TALENs. Cell Res. 2013; 23:1059–1062.

23. Sprangers B, Waer M, Billiau AD. Xenotransplantation: where are we in 2008? Kidney Int. 2008; 74:14–21.

24. Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, Jeong D, Kim JS, Lee HW. Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol. 2013; 31:23–24.

25. Tan W, Carlson DF, Lancto CA, Garbe JR, Webster DA, Hackett PB, Fahrenkrug SC. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc Natl Acad Sci U S A. 2013; 110:16526–16531.

26. Tesson L, Usal C, Ménoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, Gregory PD, Anegon I, Cost GJ. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011; 29:695–696.

27. Whitworth KM, Lee K, Benne JA, Beaton BP, Spate LD, Murphy SL, Samuel MS, Mao J, O'Gorman C, Walters EM, Murphy CN, Driver J, Mileham A, McLaren D, Wells KD, Prather RS. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol Reprod. 2014; 91:78.
28. Xin J, Yang H, Fan N, Zhao B, Ouyang Z, Liu Z, Zhao Y, Li X, Song J, Yang Y, Zou Q, Yan Q, Zeng Y, Lai L. Highly efficient generation of GGTA1 biallelic knockout inbred mini-pigs with TALENs. PLoS One. 2013; 8:e84250.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download