Abstract
Infection of cattle with bovine leukemia virus (BLV) has been observed and reported worldwide, including in Korea. The onsite identification of infected cattle would help decreasing and eradicating BLV infections on farms. Here, we present a new immunochromatographic assay that employs monoclonal antibodies (MAbs) for the detection of antibodies against BLV in the field. BLV envelope glycoprotein (gp)51 was expressed in E. coli, and MAbs against recombinant BLV gp51 were generated for the development of an immunochromatographic assay to detect BLV antibodies in cattle. The sensitivity and specificity of the assay were determined by comparing these results with those obtained from a standard enzyme linked immunosorbent assay (ELISA). A total of 160 bovine sera were used to evaluate the new immunochromatographic assay. Using ELISA as a reference standard, the relative specificity and sensitivity of this assay were determined to be 94.7% and 98%, respectively. Because of its high sensitivity and specificity, this BLV antibody detection assay would be suitable for the onsite identification of BLV infection in the field.
Bovine leukemia virus (BLV), which belongs to the genus Deltaretrovirus of the Retroviridae family, is the causative agent of enzootic bovine leukosis (EBL). This disease results in economic losses for the cattle industry worldwide [5]. BLV infections are found worldwide, but in recent years many European countries have successfully eradicated EBL [1572526273537]. Although BLV-infected cattle are asymptomatic, 30 to 70% of infected animals develop persistent lymphocytosis and 0.1 to 10% develop lymphoid tumors [1825].
The envelope (env) protein complex of BLV is composed of surface glycoprotein subunits (gp51) that are anchored to virions by their association with transmembrane protein subunits (gp30) [15]. The BLV envelope glycoproteins play a crucial role in the virus life cycle by mediating viral entry through specific interactions with cell surface receptors and establishing cellular tropism. As a result, they are also the natural targets of neutralizing antibodies [23].
Recent phylogenetic analyses of the BLV env genes showed that BLV can be classified into eight or more genotypes [232]. BLV infection was first detected in Korea in the 1980s by serology testing [6]. In recent studies, the prevalence of EBL in dairy cattle has been determined to be greater than 50% [317]. The most recent Korean BLV isolates characterized were classified as BLV genotypes 1 and 3 [20].
Previous studies have identified diverse routes of EBL transmission among cattle, and it is well known that cattle are infected with BLV through the transfer of blood and blood products that contain BLV-infected lymphocytes [30]. Routine farm practices, such as tattooing [21], dehorning [9], and rectal palpation [13], are also likely to lead to viral transmission. Insect vectors and other large biting flies have also been known to transmit the virus [3]. Additionally, vertical transmission may occur transplacentally from an infected dam to her fetus or by the newborn calf's ingestion of infected colostrum [1030].
The prevention of blood transmission between infected and naïve animals is the most important aspect of prevention protocols [122431]. Good practices recommended by the New York State Bovine Leukosis Virus Eradication and Certification Program (NYSBLVECP) in 1985 strongly suggested that infected and uninfected animals be separated to reduce close contact [4]. Infected animals can be identified by serological and molecular diagnosis [38]; however, most of the diagnostic methods available for BLV detection must be performed by trained experts in laboratories with specialized equipment, and time is required to obtain the final results. Accordingly, a point-of-care testing (POCT) method is needed to efficiently segregate BLV-infected from naive animals. Here, we describe a pilot study in which a novel, rapid immunochromatography assay combining monoclonal antibodies (MAbs) and colloidal gold particles for the detection of BLV antibodies for use as a POCT method was developed and evaluated.
Animal immunization experiments were approved by the Institutional Animal Care and Use Committee (IACUC No. MD 2014-006) of Median Diagnostics, Korea, and were carried out in strict accordance with the Care and Use of Laboratory Animals regulations of the Ministry of Agriculture, Food and Rural Affairs, Korea. BALB/c mice were purchased from Orient Bio, Korea. BLV sera for use as positive controls were obtained from the National Veterinary Services Laboratories (NVSL, USA). BLV reference serum of World Organization for Animal Health (OIE), E05 was kindly provided by Prof. Vahlenkamp of the Institute of Virology, Centre for Infectious Diseases, Faculty of Veterinary Medicine, Leipzig University, Germany. Antisera against other bovine viral diseases, such as bovine viral diarrhea virus (BVDV), bovine respiratory syncytial virus (BRSV), bovine herpesvirus-1 (BHV-1-1), and parainfluenza-3 (PI-3) virus, were also obtained from the NVSL. Field bovine serum samples (n = 160) were collected from cattle in the Southern part of Korea (Gyeongbuk province) between April and July of 2014.
A 648 bp fragment of the env gene spanning the region between nucleotide positions 100 and 747, which encodes a partial BLV gp51 totaling 216 amino acids, was amplified from genomic DNA (GenBank accession No. KP201460) [20] that had been extracted from the peripheral blood mononuclear cells of BLV-infected cattle collected in Korea in April 2014. The N-terminal portion of the gp51 protein was expressed as a 6× histidine-tagged recombinant protein using the pET-21a(+) plasmid (Novagen, USA) and E. coli strain BL21 (Yeastern Biotech, Taiwan) according to the manufacturer's protocols. Briefly, part of the gp51 coding region was amplified using the primers BLV-P3 F-5′-CGCGAATTCTGGAGATGCTCCCTG TCCCTAGGAAA-3′ and BLV-P3 R-5′-CGCCTCGAGCCAG AAGATTTGGGCGTCC-3′. Polymerase chain reaction was performed using a T3000 thermocycler (Biometra, Germany). The gp51 gene was cloned into the pET-21a(+) vector using the restriction enzymes BamHI and XhoI (New England Biolabs, UK) and subsequently transformed into BL21 cells to express the 6× His-tagged fusion proteins. After induction with 0.2 mM IPTG (Armresco, USA) for 20 h at 25℃, the bacterial cell pellet was sonicated in chromatography buffer (20 mM sodium phosphate, 500 mM sodium chloride, 8 M Urea, and 20 mM imidazole, pH 7.4) and purified using Ni-NTA agarose (Qiagen, Germany) [39]. The affinity-purified protein was dialyzed against a urea gradient prior to use. The recombinant gp51 protein was solubilized in 8 M urea buffer, which was then exchanged for 150 mM Tris-HCl (iNtRON, Korea). Purified antigens were evaluated by polyacrylamide gel electrophoresis (SDS-PAGE) followed by protein staining with Coomassie G250 (Invitrogen).
At eight weeks of age, BALB/c mice were immunized intraperitoneally with 0.2 mL (0.5 mg/mL) of recombinant partial BLV gp51 protein emulsified 1 : 1 in Freund's complete adjuvant (Sigma, USA). The mice were boosted three times with the same quantity of antigen at two week intervals. Four days after the last immunization, their splenic mononuclear cells were isolated and fused with murine myeloma cells (SP 2/0 Ag14) (ATCC, USA) using 50% polyethylene glycol (PEG)-1000 (Roche, Germany). The hybridomas were generated by HAT medium selection (Sigma) and screened using a recombinant BLV protein-based enzyme linked immunosorbent assay (ELISA). The positive hybridoma cells were cloned by limiting-dilution, and 18 positive hybridoma lines were obtained. The selected MAbs were further tested for their ability to interfere with ligand-receptor binding in a blocking ELISA, as previously described [28]. Western blot and isotype analyses were performed as previously described [34] using the hybridoma with superior blocking ability.
Indirect ELISAs were performed to screen the established hybridomas. Briefly, 96-well plates were coated with 100 µL of 0.5 µg/mL BLV gp51 protein or BLV virions in coating buffer as the coating antigens for 1 h at 37℃. After being washed three times with PBS containing 0.05% Tween 20 (PBS-T; Sigma), the wells were blocked with PBS containing 10% FBS for 1 h at 37℃. Following blocking, 100 µL of supernatant from the hybridoma cultures serially diluted in ELISA buffer (PBS containing 0.5% bovine serum albumin [BSA] and 0.1% Tween 20; Sigma) was added to the wells. After incubation for 1 h at room temperature, the wells were washed three times with PBS-T. Bound antibodies were then detected with horseradish peroxidase (HRP)-labeled goat anti-mouse IgG antibody (KPL, USA). Following washing, the colorimetric reaction was measured using 3,3',5,5'-tetramethylbenzidine (TMB) peroxidase substrate (KPL) after a 10-min development period. The reaction was then terminated with 1 M sulfuric acid (Sigma) and the absorbance at 450 nm was measured using a spectrophotometer (Tecan, Switzerland).
Blocking ELISA for the additional screening of MAbs with blocking ability was carried out as previously described [28]. Briefly, plates were coated with 100 µL of BLV (1 µg/mL) that had been concentrated and purified as previously described [14]. After three washes with PBS-T, the wells were blocked with PBS containing 10% FBS for 1 h at 37℃. Following blocking, 100 µL of BLV antiserum diluted 1 : 1 in ELISA buffer (PBS containing 0.1% Tween 20) were added to the wells and incubated for 1 h at 37℃. After washing, the MAbs were diluted 1 : 10 in PBS, added to the plates (50 µL/well) and incubated for 1 h at 37℃. After further washing, bound MAbs were detected by incubation with HRP-labeled goat anti-mouse IgG antibodies for 1 h at 37℃. The colorimetric reaction was measured by treating with TMB substrate and allowing the reaction to develop for 10 min. The reaction was terminated with 1 M sulfuric acid and the absorbance at 450 nm was measured.
Colloidal gold was prepared as previously described [11], with minor modifications. Briefly, 1 mL of a 1% solution of chloroauric acid (Sigma) was added to 100 mL of distilled water and boiled. While boiling, 2.4 mL of a 1% solution of trisodium citrate dihydrate was added to the solution. The gold solution gradually formed as the citrate reduced the gold (III). When the solution began to develop a deep red color, heating was discontinued. Colloidal gold (40 nm) (Median Diagnostics) and BLV lysate antigen were prepared as previously described [28] and added drop-wise into the colloidal gold solution (final concentration 10 µg/mL) over a 1 h period at room temperature while being stirred. Casein (Sigma) was added as a blocking agent (final concentration at 0.3%), and the mixture was stirred for 1 h at room temperature. After the mixture was centrifuged at 14,000 × g and 4℃ for 30 min, the supernatant was discarded and the resulting pellet was resuspended in 1/10 of its original volume in 2 mM borax buffer containing 1% BSA. Following removal of the colloidal gold-recombinant antigen conjugate to a 1.5 mL tube, 5% sucrose (Sigma), 0.1% casein, and 0.1% sodium azide (Sigma) were added, and the concentration of the conjugate was adjusted to OD450 = 1.5. Following these steps, the conjugate solution was allowed to dry.
The immunochromatographic strip consists of a sample pad, a nitrocellulose membrane, and an absorbance pad mounted on a plastic support. MAb 12F12 and anti-rabbit IgG (Arista Biologicals, USA) were diluted with 0.5% sucrose (Sigma) in 2.5 mM PBS (pH 7.0) to obtain final concentrations of 1 mg/mL and 2 mg/mL, respectively. Anti-rabbit IgG was then immobilized in the control line position and MAb 12F12 was immobilized in the test line position on a nitrocellulose membrane (Millipore, Germany) attached to a backing card (PJ Company, Korea). After the membrane was dried in a dehumidification chamber for more than 24 h, it was cut into 4 mm wide strips using a cutting machine (Zeta Corporation, Korea). Each membrane piece was then assembled on a membrane cassette with a sample pad and an absorbance pad. The strips were then stored under dry conditions at room temperature until used.
During testing, serum samples reacted with the colloidal gold-recombinant antigen conjugate. After the sample-conjugate mixture was added to the specified sample application area on the immunochromatographic strip, the blocked antigen would not react with the BLV-specific MAb 12F12 that had been immobilized on the nitrocellulose membrane, and the test line would not indicate a positive result by developing any gold color. A brief description of the test procedure is as follows. The serum dilution was prepared by adding three drops (90 µL) of sample buffer to the dried antigen-gold conjugate with a dropper, followed by the addition of three drops of test serum. After these components were mixed well, three drops of the mixture were added to the strip sample application area. Following incubation for 10 min at room temperature, the line in the strip was examined by the naked eye or by a Time-Resolved Fluorescence detector (Medisensor, Korea). A serum sample that resulted in the appearance of both test and control lines was considered negative, whereas a serum sample that only resulted in the development of the control line was considered positive (Fig. 1). A Time-Resolved Fluorescence detector value of < 200 of the test line indicated a positive result.
Correlations between the results of the immunochromatographic strip assay and a reference method were evaluated using 160 bovine serum samples. A commercially available ELISA kit, the BLV gp51-Ab kit (IDEXX Laboratories, France), was selected as the reference test for comparison and utilized according to the manufacturer's protocol.
The efficacy of the immunochromatographic assay was compared to that of the ELISA using 160 field sera samples. The kappa value [19] was estimated as a measure of agreement between the two tests. Kappa values greater than 0.75 were considered to represent agreement between test results.
The partial BLV gp51 protein was expressed in E. coli. Following purification, the presence of the 27.5 kDa recombinant antigens was confirmed by SDS-PAGE and protein staining (Fig. 2).
Hybridomas were screened using indirect ELISAs with both the purified recombinant gp51 protein and BLV particles. An absorbance < 0.2 was considered negative. Among the clones tested, 18 hybridomas produced a positive signal and were selected for further analysis (Table 1). These hybridomas were evaluated using a blocking ELISA with BLV and anti-BLV serum to identify MAbs that demonstrated blocking ability. Among the hybridomas tested, seven (8A8, 10B43, 9D72, 2E42, 8F76, 12F12, 10H37) exhibited blocking ability (Fig. 3). The reactivity of the selected MAbs against BLV gp51 was further studied by examining their interaction with BLV-gold conjugates embedded in nitrocellulose membranes. Following these tests, MAb 12F12 was selected for assay development after exhibiting strong reactivity (Fig. 4).
The specificity of the immunochromatographic assay strip was evaluated using antisera targeting a range of bovine viral agents (BVDV, BRSV, BHV-1-1, PI-3 and BLV). Each antiserum was treated as specified in the Materials and Methods and applied to the strip. The procedure was performed in triplicate for each antiserum. BLV antiserum resulted in one strong band on the control line, whereas the remaining samples produced clear bands on both the control and sample lines (Fig. 5). Importantly, the BLV reference serum E05 diluted 1 : 10 showed a positive result in the immunochromatographic assay (Fig. 6).
Of 160 bovine field sera examined, 57 samples tested positive for BLV using a commercially available bovine leukosis blocking ELISA kit (IDEXX Laboratories). These same serum samples were tested using the newly developed immunochromatographic assay. Of the 160 samples, 56 were positive. Thus, the specificity and sensitivity of the immunochromatographic assay were found to be 94.7% and 98%, respectively, when compared to the ELISA results (Table 2). Moreover, there was good agreement (κ = 0.932) between the results of the two tests.
The reproducibility of the assay strip was determined by comparing the results generated by strips from several different batches. Two BLV antibody-positive samples (one from NVSL and one from the field) and one negative serum sample were used, and the test was performed in triplicate. The results indicated that there was limited variation among the three repeated tests (Table 3; Fig. 7), and the coefficient of variation was less than 10%.
BLV exists as an integrated proviral DNA in host cells [18] and thus causes lifelong infection in cattle [38]. In cattle, antibodies against BLV can first be detected 3 to 16 weeks after infection; therefore, detection of these antibodies is one of the methods used to identify EBL [38]. The immune response to BLV infection includes the constitutive production of antibodies targeting the BLV gp51 protein [2]. These antibodies are present in higher titers and appear earlier than other BLV-specific antibodies raised against the major internal viral protein, p24 [22]. This expression pattern explains why most serological assays for EBL, such as ELISA, were developed using gp51 or whole BLV as the target [829].
In this study, an immunochromatographic assay using a MAb against the BLV gp51 antigen was developed to detect antibodies against BLV in the field. A truncated gp51 protein of 27.5 kDa (216 amino acids) was expressed in E. coli, and MAbs against the recombinant gp51 were produced. The reactivity of the generated hybridomas was tested using indirect ELISA with the recombinant gp51 protein as the target antigen, and 18 hybridomas tested positive and were selected. Among these selected hybridomas, seven MAbs with blocking activity were identified using blocking ELISA. The MAb from hybridoma 12F12 was finally selected for assay development because it reacted strongly with the BLV-gold conjugate on the nitrocellulose membrane.
The BLV lysate was conjugated to colloidal gold, after which the conjugate was aliquoted into 1.5 mL tubes and allowed to dry. Bovine serum samples were applied to the recombinant env protein-gold conjugate, and BLV antibodies in positive samples were expected to bind to the recombinant env in this mixture. This sample solution was then applied to a nitrocellulose membrane that contained an immobilized MAb against the N terminal gp51 protein fragment. Any unbound recombinant proteins conjugated to the gold in the diluted samples that were negative for the BLV antibody would bind to the MAb on the nitrocellulose membrane and cause a colorimetric reaction.
The immunochromatographic assay was determined to be 94.7% specific and 98% sensitive compared to the commercial BLV ELISA kit. Moreover, a kappa value of 0.93 indicated excellent agreement between the results of these two tests. OIE reference sera for EBL diagnosis, E05 are available from the OIE Reference Laboratory. In assays in which serum samples are tested individually, OIE reference serum E05 diluted 1 : 10 must be positive (OIE Terrestrial Manual) [38]. The 1 : 10-diluted E05 BLV reference serum tested positive in this immunochromatographic assay. Additionally, there was no specific binding detected when antibodies targeting other bovine viral agents (BVD, BRSV, BHV-1, or PI-3) were assessed using this immunochromatographic assay.
This report describes the development of the first POCT assay for the detection of BLV antibodies using a blocking format. This methodology will be useful in areas in which the EBL eradication policy is practiced, as well as in countries where EBL is still present and efforts are being made to reduce its overall prevalence. Furthermore, the assay will be valuable in countries in which laboratory tests are not readily available because it will allow suspected animals to be segregated from uninfected animals prior to laboratory confirmation.
Figures and Tables
Fig. 1
Interpretation of the immunochromatographic assay results. (A) A serum sample that only produced the control line was considered positive. (B) A serum sample that produced both the test and control lines was considered negative.

Fig. 2
SDS-PAGE analysis of the recombinant bovine leukemia virus (BLV) gp51 protein. (A) Before purification. Lane 1, protein size marker; Lane 2, BL21(DE3) total cell lysate; Lane 3, BL21(DE3) cell lysate supernatant; Lane 4, recombinant gp51 protein in BL21(DE3) total cell lysate; Lane 5, recombinant gp51 protein in BL21(DE3) cell lysate supernatant. (B) After purification. M, marker; T, BL21(DE3) total cell lysate; S, BL21(DE3) cell lysate supernatant; F, flow through; 40, flow through after wash step; 60, flow through after wash step; E, eluted gp51 protein.
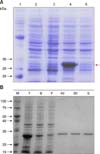
Fig. 3
Blocking ELISA using BLV and BLV antiserum for the selection of monoclonal antibodies (MAbs) with blocking ability. Of the 18 hybridoma clones that were tested, seven (hybridoma clones 8A8, 10B43, 9D72, 2E42, 8F76, 12F12, and 10H37) exhibited superior blocking ability.

Fig. 4
Reactivity of the selected MAbs against BLV gp51 in the BLV-gold conjugate immobilized on nitrocellulose membranes. Of the seven hybridoma clones tested, hybridoma clone 12F12 was selected, as it reacted strongly with BLV in the immunochromatography strip assay.

Fig. 5
Specificity of the immunochromatographic assay strip. The specificity of the immunochromatographic assay strip was evaluated with antisera against a range of bovine viral agents, including BVDV, BRSV, BHV-1-1, PI-3 and BLV. *Value read in the reader.

Fig. 6
Evaluation of the immunochromatographic assay strip using the BLV reference serum E05 diluted 1 : 10 in serum negative for BLV antibody. Lane 1, E05 serum diluted 1 : 10; Lane 2, BLV-antibody-negative serum 1; Lane 3, serum that was positive for the BLV antibody (NVSL, USA); Lane 4, BLV-antibody-negative serum 2.

Fig. 7
Reproducibility of the immunochromatographic assay strip. Sera that were positive and negative for the BLV antibody were tested in triplicate. Lane 1, serum that was positive for the BLV antibody (NVSL); Lane 2, field-collected bovine serum that was positive for the BLV antibody (as determined by ELISA); Lane 3, serum that was negative for the BLV antibody.

Table 1
Hybridomas screened by indirect enzyme linked immunosorbent assay (ELISA) and immunofluorescence assay

Acknowledgments
This study was funded by the grant from the Ministry of Agriculture, Food and Rural Affairs, Republic of Korea to the Animal and Plant Quarantine Agency.
References
1. Acaite J, Tamosiunas V, Lukauskas K, Milius J, Pieskus J. The eradication experience of enzootic bovine leukosis from Lithuania. Prev Vet Med. 2007; 82:83–89.

2. Balić D, Lojkić I, Periškić M, Bedeković T, Jungić A, Lemo N, Roić B, Cać Z, Barbić L, Madić J. Identification of a new genotype of bovine leukemia virus. Arch Virol. 2012; 157:1281–1290.

3. Bech-Nielsen S, Piper CE, Ferrer JF. Natural mode of transmission of the bovine leukemia virus: role of bloodsucking insects. Am J Vet Res. 1978; 39:1089–1092.
4. Brunner MA, Lein DH, Dubovi EJ. Experiences with the New York State Bovine Leukosis Virus Eradication and Certification Program. Vet Clin North Am Food Anim Pract. 1997; 13:143–150.

5. Burny A, Cleuter Y, Kettmann R, Mammerickx M, Marbaix G, Portetelle D, van den, Willems L, Thomas R. Bovine leukaemia: facts and hypotheses derived from the study of an infectious cancer. Vet Microbiol. 1988; 17:197–218.

6. Choi WP. Survey for antibodies to bovine leukemia virus in dairy and Korean native cattle. Korean J Vet Res. 1982; 22:23–26.
7. D'Angelino JL, Garcia M, Birgel EH. Epidemiological study of enzootic bovine leukosis in Brazil. Trop Anim Health Prod. 1998; 30:13–15.
8. De Giuseppe A, Feliziani F, Rutili D, De Mia GM. Expression of the bovine leukemia virus envelope glycoprotein (gp51) by recombinant baculovirus and its use in an enzyme-linked immunosorbent assay. Clin Diagn Lab Immunol. 2004; 11:147–151.

9. DiGiacomo RF, Darlington RL, Evermann JF. Natural transmission of bovine leukemia virus in dairy calves by dehorning. Can J Comp Med. 1985; 49:340–342.
10. Dimmock CK, Chung YS, MacKenzie AR. Factors affecting the natural transmission of bovine leukaemia virus infection in Queensland dairy herds. Aust Vet J. 1991; 68:230–233.

11. Frens G. Controlled nucleation for the regulation of the particle size in monodisperse gold suspensions. Nat Phys Sci. 1973; 241:20–22.

12. Gutiérrez G, Alvarez I, Politzki R, Lomónaco M, Dus Santos MJ, Rondelli F, Fondevila N, Trono K. Natural progression of bovine leukemia virus infection in Argentinean dairy cattle. Vet Microbiol. 2011; 151:255–263.

13. Hopkins SG, DiGiacomo RF, Evermann JF, Christensen JD, Deitelhoff DP, Mickelsen WD. Rectal palpation and transmission of bovine leukemia virus in dairy cattle. J Am Vet Med Assoc. 1991; 199:1035–1038.
14. Jafari Jozani R, Taheri M, Khazraiinia P, Hemmatzadeh F. Using various antigen preparations to produce monoclonal antibodies against bovine leukaemia virus (BLV) gp51SU. Iran J Vet Res. 2008; 9:25–31.
15. Johnston ER, Albritton LM, Radke K. Envelope proteins containing single amino acid substitutions support a structural model of the receptor-binding domain of bovine leukemia virus surface protein. J Virol. 2002; 76:10861–10872.

16. Jun MH, Chung UI, An SH. Studies on bovine lymphosarcoma. III. Preparation of bovine leucosis virus antigen and its biological properties. Res Rep ORD. 1983; 25:68–74.
17. Jung K, Shim HS, Baek JJ. Investigation of bovine leukemia virus infection in dairy farms of northern Gyeonggi province, Korea. Korean J Vet Ser. 2012; 35:333–337.

18. Kettmann R, Cleuter Y, Mammerickx M, Meunier-Rotival M, Bernardi G, Burny A, Chantrenne H. Genomic integration of bovine leukemia provirus: comparison of persistent lymphocytosis with lymph node tumor form of enzootic. Proc Natl Acad Sci U S A. 1980; 77:2577–2581.
19. Landis JR, Koch GG. An application of hierarchical kappa-type statistics in the assessment of majority agreement among multiple observers. Biometrics. 1977; 33:363–374.

20. Lee E, Kim EJ, Joung HK, Kim BH, Song JY, Cho IS, Lee KK, Shin YK. Sequencing and phylogenetic analysis of the gp51 gene from Korean bovine leukemia virus isolates. Virol J. 2015; 12:64.

21. Lucas MH, Roberts DH, Wibberley G. Ear tattooing as a method of spread of bovine leukosis virus infection. Br Vet J. 1985; 141:647–649.

22. Mammerickx M, Portetelle D, Burny A, Leunen J. Detection by immunodiffusion- and radioimmunoassay-tests of antibodies to bovine leukemia virus antigens in sera of experimentally infected sheep and cattle. Zentralbl Veterinarmed B. 1980; 27:291–303.

23. Mamoun RZ, Morisson M, Rebeyrotte N, Busetta B, Couez D, Kettmann R, Hospital M, Guillemain B. Sequence variability of bovine leukemia virus env gene and its relevance to the structure and antigenicity of the glycoproteins. J Virol. 1990; 64:4180–4188.

24. Maresca C, Costarelli S, Dettori A, Felici A, Iscaro C, Feliziani F. Enzootic bovine leukosis: report of eradication and surveillance measures in Italy over an 8-year period (2005-2012). Prev Vet Med. 2015; 119:222–226.

25. Mirsky ML, Olmstead CA, Da Y, Lewin HA. The prevalence of proviral bovine leukemia virus in peripheral blood mononuclear cells at two subclinical stages of infection. J Virol. 1996; 70:2178–2183.

26. Murakami K, Kobayashi S, Konishi M, Kameyama K, Yamamoto T, Tsutsui T. The recent prevalence of bovine leukemia virus (BLV) infection among Japanese cattle. Vet Microbiol. 2011; 148:84–88.

27. Nuotio L, Rusanen H, Sihvonen L, Neuvonen E. Eradication of enzootic bovine leukosis from Finland. Prev Vet Med. 2003; 59:43–49.

28. Oem JK, Chang BS, Joo HD, Yang MY, Kim GJ, Park JY, Ko YJ, Kim YJ, Park JH, Joo YS. Development of an epitope-blocking-enzyme-linked immunosorbent assay to differentiate between animals infected with and vaccinated against foot-and-mouth disease virus. J Virol Methods. 2007; 142:174–181.

29. Portetelle D, Mammerickx M, Burny A. Use of two monoclonal antibodies in an ELISA test for the detection of antibodies to bovine leukaemia virus envelope protein gp51. J Virol Methods. 1989; 23:211–222.

30. Reed VI. Enzootic bovine leukosis. Can Vet J. 1981; 22:95–102.
31. Rodríguez SM, Florins A, Gillet N, de Brogniez A, Sánchez-Alcaraz MT, Boxus M, Boulanger F, Gutiérrez G, Trono K, Alvarez I, Vagnoni L, Willems L. Preventive and therapeutic strategies for bovine leukemia virus: lessons for HTLV. Viruses. 2011; 3:1210–1248.

32. Rola-Łuszczak M, Pluta A, Olech M, Donnik I, Petropavlovskiy M, Gerilovych A, Vinogradova I, Choudhury B, Kuźmak J. The molecular characterization of bovine leukaemia virus isolates from Eastern Europe and Siberia and its impact on phylogeny. PLoS One. 2013; 8:e58705.

33. Suh GH, Lee CG, Lee CY, Hur TY, Kang SJ, Son DS, Ryu IS, Ahn B, Kim NC, Joo YS. Prevalence of anti-bovine leukemia virus antibodies in dairy and Korean native cattle. J Vet Clin. 2003; 20:172–176.
34. Troiano LD, Thomaz-Soccol V, Agottani JVB, Brodzinski J, Penha TR, Ozaki SC. Production, characterization, and use of monoclonal antibodies against gp51 protein to diagnose bovine leukemia virus infection. Biores Open Access. 2013; 2:55–60.

35. Trono KG, Pérez-Filgueira DM, Duffy S, Borca MV, Carrillo C. Seroprevalence of bovine leukemia virus in dairy cattle in Argentina: comparison of sensitivity and specificity of different detection methods. Vet Microbiol. 2001; 83:235–248.

36. Van Der Maaten MJ, Miller JM. Replication of bovine leukemia virus in monolayer cell cultures. Bibl Haematol. 1975; 360–362.

37. VanLeeuwen JA, Keefe GP, Tremblay R, Power C, Wichtel JJ. Seroprevalence of infection with Mycobacterium avium subspecies paratuberculosis, bovine leukemia virus, and bovine viral diarrhea virus in maritime Canada dairy cattle. Can Vet J. 2001; 42:193–198.
38. World Organisation for Animal Health. Chapt. 2.4.11. Enzootic bovine leukosis. Manual of Diagnositic Tests and Vaccines for Terresrtial Animals. Paris: OIE;2015. p. 721–732.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download