Abstract
Drug-induced liver injury (DILI) is a significant threat to patient health and a major concern during drug development. Recently, multiple circulating microRNAs (miRNAs) have been reported to be potential biomarkers for DILI. To adapt and validate miRNAs for clinical use, we investigated the time-course changes in miR-122 expression levels in an acetaminophen-induced liver injury model in rats. In addition, miR-155 and miR-21 were evaluated as makers of inflammation and regeneration, respectively, to characterize liver status. Our results revealed that miR-122 is an early and sensitive biomarker of hepatocellular injury at a stage when alanine transaminase, aspartate transaminase, and total bilirubin were not detectable. However, no significant differences in the expression levels of other miRNAs (miR-155 and -21) were observed between treatment and vehicle groups. Collectively, these time-course changes in the expression levels of miRNAs may be useful as markers for clinical decision-making, in the diagnosis and treatment of DILI.
Drug-induced liver injury (DILI) is a serious clinical problem and the leading cause of death from acute liver failure, accounting for approximately 13% of all cases of acute liver failure in the United States [10]. Furthermore, drug-induced hepatotoxicity is a major concern for post-marketing regulatory decisions, including drug withdrawal [20]. Although alanine transaminase (ALT) is a relatively sensitive biomarker of liver injury, it does not always correlate with adverse effects in the human liver [4].
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression by binding to mRNAs and interfering with the process of translation [2]. Two liver-enriched miRNAs (miR-122 and miR-192) are promising serum biomarkers of acetaminophen (N-acetyl-p-aminophenol; APAP)-induced acute liver injury in mice [24]. Furthermore, recent evidence suggests that miRNAs are potential biomarkers of acute liver injury in humans [32226]. Recently, miR-155, a master regulator of inflammation, was shown to be up-regulated in infl ammation-related diseases in mice [7]. miR-21 also regulates various genes involved in the cell cycle and DNA synthesis and is a potential biomarker of regeneration in the liver [13]. Hepatocellular injury, inflammation, and regeneration can differ between liver diseases of various etiologies, and these miRNAs must be validated to adapt them for clinical use.
In acute overdose of APAP, N-acetyl-p-benzoquinone-imine, a toxic metabolite and highly reactive electrophile, is generated [24]. N-acetylcysteine (NAC) can be quickly converted into intracellular GSH in the body [5] and effectively prevent and reduce APAP-induced liver injury if administered within 8–10 h of initial ingestion [11]. However, APAP overdose does not usually present immediate symptoms during the first 24 h of ingestion [24], and adverse reactions to NAC, such as vomiting, nausea, and anaphylactoid reaction, are common. Therefore, careful patient selection is essential, and early and sensitive blood-based biomarkers for APAP overdose are urgently required.
This study was conducted to evaluate changes in the time-course of expression levels of miRNAs in acute liver injury models. To adapt miRNAs as liver-specific biomarkers in humans, it is important to discriminate between transient damage/repair and progressive liver injury to determine whether serum miR-122 has clinical prognostic value over current biomarkers of APAP-induced liver injury. In addition, use of specific miRNA biomarkers (miR-122, -155, and -21) can enable evaluation of correlations in the expression and histopathology of liver cell damage, inflammation, and regeneration in the context of acute liver injury. To accomplish this, we examined the expression levels of miRNAs at multiple time points (0, 3, 6, 24, 48, and 72 h after dosing) in an APAP-induced liver injury model in rats. All plasma miRNA levels were examined by correlation with histopathology and other serum biomarkers (ALT, aspartate transaminase [AST], and total bilirubin) to validate the usefulness of miRNAs as serum biomarkers.
A total of 36 healthy, approximately 6-week-old, male Sprague-Dawley rats were supplied by Orient Bio (Korea) and acclimatized for 2 weeks before the start of the experiment. Animals were group housed (3–4 rats/cage) in polysulfone cages with a wire mesh roof under routine conditions of temperature, relative humidity, ventilation, and illumination. Rats were fed an autoclaved pellet diet (PMI certified Rodent LabDiet No. 5002; Land O'Lakes, USA) ad libitum with free and continuous access to drinking water. Prior to blood sampling and euthanasia, animals had access to water and were not fasted. The study protocol was reviewed and approved by the Animal Care and Use Committee of the Asan Medical Center (IACUC No. 2014-14-072).
The study design is depicted in Fig. 1. Prior to the start of treatment, rats were randomly allocated to the vehicle (one group of six rats) or APAP-dosed groups (five groups of six rats each). Rats were treated with a single dose of APAP (1,000 mg/kg orally; Sigma, USA) or vehicle (0.5% carboxy methylcellulose; Sigma). The level of APAP was set at the maximum tolerated dose based on clear signs of toxicity with little or no lethality [121]. The injection volume for each treatment or vehicle was 10 mL/kg body weight. Clinical observations were performed daily, and body weight was recorded prior to dosing and euthanasia. Blood was collected from the caudal vena cava before necropsy, which was performed 6 h after dosing in the control group and 3, 6, 24, 48, or 72 h after dosing in the treatment group after individual and humane euthanasia.
Serum ALT, AST, and total bilirubin levels were measured using an automated clinical chemistry analyzer (Hitachi 7180; Hitachi High-Technologies, Japan).
EDTA was added to blood as an anticoagulant, and plasma was collected after centrifugation. Total RNA (including small RNA) was prepared using a NucleoSpin miRNA kit (Macherey-Nagel, Germany). cDNA was synthesized from total RNA using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, USA). Reverse transcription was carried out in a total volume of 15 µL with reagents from the TaqMan MicroRNA RT Kit (Applied Biosystems). Briefly, 5 µL of purified total RNA was added to 7 µL of master mix (3 µL of 5× multiplex RT primer pool, 0.15 µL of 100 mM deoxynucleotide triphosphates (dNTPs), 1 µL of 50 U/µL MultiScribe reverse transcriptase, 1.5 µL of 10× RT buffer, 0.19 µL of RNase inhibitor, and 4.16 µL of RNase-free water). The reaction was mixed and incubated on ice for 5 minutes after which it was subjected to 30 min at 16℃, 30 min at 42℃, and 5 min at 85℃, followed by incubation at 4℃.
Real-time PCR assays of individual miRNAs were conducted in a total volume of 20 µL [1 µL of 20× TaqMan primer, 5 µL of 2× TaqMan Universal PCR Master Mix (Applied Biosystems), 7.67 µL of RNase-free water, and 1.33 µL of diluted cDNA]. Real-time PCR was performed in duplicate on a 7900 HT Fast Real-Time PCR System (Applied Biosystems). U6 was used as a reference gene for normalization. The sequences of purchased miRNA probes were as follows: miR-122 (rno-miR-122-5p, rat, UGGAGUGUGACAAUGGUGUUUG, Applied Biosystems), miR-155 (rno-miR-155-5p, rat, UUAAUGCUAAUUGUGAUAGGGGU, Applied Biosystems), miR-21 (rno-miR-21-5p, rat, UAGCUUAUCAGACUGAUGUUGA, Applied Biosystems). For each sample in all treatment groups, the fold change relative to the median vehicle sample was calculated using the comparative Ct method [15].
Necropsy was performed on all animals, with macroscopic evaluation of the thoracic and abdominal cavity and tissues. After macroscopic examination, the excised liver was fixed in 10% neutral buffered formalin. Two lobes (the left lateral lobe and the median lobe) were sectioned and washed with tap water for approximately 6 h to remove formaldehyde. Tissues were then dehydrated in graded ethanol and cleared in xylene using an Excelsior S tissue processor (Thermo Scientific, USA), after which they were embedded in paraffin blocks using an EG1150H paraffin embedding station (Leica, Germany). The paraffin blocks were cut into 4 µm sections on an RM2255 rotary microtome (Leica).
Sections were mounted onto glass slides, and hematoxylin and eosin staining and coverslipping were performed on an Autostainer XL (Leica). Histological examination of livers was performed by an experienced pathologist. Centrilobular necrosis with inflammatory cell infiltration was graded as follows: 0, normal; 1, minimal; 2, slight; 3, moderate; 4, marked; and 5, severe.
Data are expressed as the means ± SEM. Significance differences between the APAP-dosed and vehicle groups were assessed by ANOVA and Dunnett's post hoc test using GraphPad Prism (GraphPad Software, USA). Fold changes in biomarker expression levels are expressed versus the vehicle group. P values < 0.05 were considered statistically significant.
Rats were treated with a single dose of APAP (1,000 mg/kg orally) or vehicle and euthanized 3, 6, 24, 48, or 72 h later. The histopathological features of APAP-treated rats were minimal to moderate centrilobular single-cell necrosis and/or apoptosis with inflammatory cell infiltrations. Typical photomicrographs of liver histopathological sections are presented in Fig. 2, and histopathologic grading of the liver is shown in Fig. 3. In the 3 h and 6 h groups, liver histology was normal and similar to that of the vehicle group. Minimal to slight centrilobular necrosis were initially observed in the 24 h group, and the lesions were most severe in the 48 h group. In the 72 h group, hepatocyte mitosis was frequently observed, with these lesions being less severe than in the 48 h group. These results indicate that histopathological changes in the liver and development of lesions were detectable at 24 h after APAP overdose, then diminished by 72 h.
Circulating miR-122 levels were measured along with ALT, AST, and total bilirubin levels (Fig. 4). Although histological evidence of necrosis was not present in the 3 h and 6 h groups, miR-122 levels were elevated (individual fold changes of 0–23) in these groups. Among the APAP-treated groups (3, 6, 24, 48, and 72 h), fold changes were highest in the 3 h group, and morphological changes were detected at later time points, suggesting that miR-122 is an early biomarker of hepatocellular injury. The fold change in miR-122 expression was slightly lower in the 72 h group, reaching a level similar to that in the vehicle group. These time-course changes in miR-122 expression place limits on the usefulness of miR-122 as a marker of late-stage APAP-induced liver status. Overall, these data suggest that the expression profile of this miRNA signature could serve as a biomarker of APAP-induced liver injury.
Although miR-122 expression was clearly elevated in the treatment groups, ALT and AST levels remained normal or were only minimally elevated (Fig. 5). Although ALT and AST are routinely used to assess liver damage after APAP overdose, in this study ALT, AST and total bilirubin (TBIL) levels were not significantly higher in the treatment groups.
In addition to miR-122, the expression levels of the inflammation marker miR-155 and the regeneration marker miR-21 were also examined (Fig. 6). Because APAP administration induces necrosis that can be accompanied by inflammation, we assessed the role of the inflammatory miRNA, miR-155. Histopathology revealed slight to moderate infiltration of centrilobular inflammatory cells in the 24 h, 48 h, and 72 h groups. In contrast, miR-155 levels in the treatment groups were similar to those of the vehicle group. At 72 h after APAP dosing, evidence of hepatocyte mitosis and regenerative hepatocytes were revealed by histology, but miR-21 levels had not changed significantly. These results indicate that miR-155 and miR-21 are not adequate markers of inflammation or regeneration in an APAP-induced hepatocellular injury model in rats.
There is an urgent need to identify and validate biomarkers with improved sensitivity and hepatic specificity to assist in the clinical management of APAP overdose [3]. Because the time of ingestion is often not accurately known, and APAP blood levels usually peak during the first few hours after ingestion, a low serum APAP level does not rule out a high level of APAP exposure [24]. Delayed presentation time, staggered overdose, and a lack of sensitivity of current clinical chemistry parameters remain critical impediments to the treatment of APAP overdose [812]. In this study, various time points (3, 6, 24, 48, and 72 h) were assessed, and miRNA levels were validated and correlated with histopathological examination of the liver in all of these time points. We confirmed that miR-122 is an early and sensitive marker of hepatocellular injury at a stage when no significant elevation of ALT, AST, or TBIL is detectable. Furthermore, adaptation of miR-122 was observed in the 72 h group, and this time-course change in miRNA expression, along with histopathological findings in the liver, are useful for the detection of DILI. However, the expression levels of other miRNAs (miR-155 and -21) that reflect inflammation and regeneration of hepatocytes did not differ significantly between the treatment and vehicle groups.
miR-122 is a sensitive circulating biomarker of liver injury. In mice, miR-122 elevation precedes the detection of ALT [24]. In this study, the expression level of miR-122 initially increased at 3 h and 6 h post-dosing, when histopathological changes were not detected. These results clearly demonstrate the usefulness of miR-122 as an early diagnostic marker of DILI. Because NAC can effectively reduce DILI within 8 to 10 h of the initial ingestion of APAP [11], the early predictive value of miR-122 may aid in the diagnosis and treatment of DILI.
Although miR-122 was differentially expressed post exposure, ALT, AST, and TBIL levels did not change or increased only minimally, despite histological evidence of necrosis. We confirmed that the changes in plasma miRNA following hepatocellular injury were more dynamic than those for ALT. Therefore, miR-122 may serve as a sensitive and specific marker, whereas changes in ALT expression are more subtle.
The overall time-course magnitude of histopathology did not correspond to elevation of miR-122 expression levels. In the 72 h group, the miR-122 level was similar to that in the vehicle group, although centrilobular necrosis with inflammation was observed by histology. The most severe grade of histopathology was observed in the 48 h group, with miR-122 peaking at 3 h. Therefore, small increases in miR-122 level do not always reflect the status of liver injury, indicating that the time of ingestion is important for the clinical diagnosis of DILI.
Moreover, the increases in miR-122 and ALT levels were lower than those reported in previous studies. Specifically, two previous reports measured miR-122 levels using a rat model of orally administered APAP 1,000 mg/kg, and fold changes of miR-122 in the 6 h group were about 5 and 50 [2125]. In addition, ALT and AST levels showed maximum fold changes compared to the control of 1.9 and 1.6, respectively, which were smaller than the previously reported values (2.2 and 2.8, respectively) [21], but correlated with the magnitude of the increase in miR-122 level.
Previous data showed that miR-155 levels increase in Kupffer cells after alcohol feeding, and that TNF is a target of miR-155 that promotes liver inflammation [14]. In addition, another study showed that increases in circulating miR-155 levels are correlated with liver inflammation [7]. The extent of hepatocyte damage and inflammation can differ among liver diseases of various etiologies and may change over the course of chronic liver disease. In an effort to adapt the inflammation marker to the DILI model, we measured the levels of miR-155. No significant differences were observed between the treatment and control groups, although slight to moderate centrilobular inflammatory cell infiltration was observed in the 24 h and 48 h groups. However, miR-155 is up-regulated in inflammation-related diseases such as cancer, autoimmune inflammation, and alcoholic liver disease [61723]. To distinguish DILI from other diseases such as biliary abnormality, viral hepatitis, hemodynamic injury, and malignancy, additional studies are needed to adapt the miR-155 as a marker for clinical use.
Various miRNAs regulate hepatocyte proliferation, and miR-21 regulates various genes involved in cell cycle and DNA synthesis [13]. Several studies have reported the induction of miR-21 during the first 24 h of liver regeneration following partial hepatectomy, making miR-21 the miRNA most consistently altered during the early stages of regeneration [91619]. In this study, time-course changes in miR-21 expression were not observed. Although massive hepatocyte regeneration occurred after partial hepatectomy in other studies, the magnitude of regeneration was low in our APAP-induced liver injury model. In this context, miR-21 was not sensitive enough to detect hepatocyte regeneration. In addition, a previous study reported inconsistent data regarding the role of miRNAs in liver regeneration [18]. Hence, further studies are needed to resolve this issue.
In summary, the time-course changes in miR-122 and serum chemistry marker expression indicate that this miRNA can be exploited as an accessible circulating biomarker of DILI, and that it could serve as an earlier and more dynamic marker than ALT. Because acute hepatic injury has many causes, including viral hepatitis and ischemic hepatic injury, accurate diagnosis of DILI is an important challenge for physicians. During clinical diagnosis and treatment, patients visit hospitals at various time points after DILI occurs, and some have ingested APAP at an unknown time or chronically. Hence, our work helps to establish a novel miRNA profile that could be used for prediction of time-dependent changes in miRNA.
In contrast, miR-155 and -21, which serve as markers of general inflammation and regeneration, respectively, did not change in our APAP-induced DILI models, even though inflammation and regeneration were revealed by histological analysis of the liver. Hence, to create miRNA profiles with clinical utility and prognostic value for DILI patients, additional studies are required to determine whether miR-122 works alone or in combination with other miRNAs.
Figures and Tables
 | Fig. 1Study design. Rats were treated with a single dose of acetaminophen (APAP; 1,000 mg/kg orally) or vehicle. Necropsy of animals in the vehicle group was performed 6 h after dosing, whereas animals in the treatment group were individually sacrificed at 3, 6, 24, 48, or 72 h after dosing. |
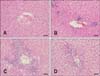 | Fig. 2Histological features of liver sections. APAP-treated rats exhibited minimal to moderate centrilobular single-cell necrosis and/or apoptosis with inflammatory cell infiltration. (A) In the 6 h group, the histology of the liver was normal. (B) In the 24 h group, minimal to slight centrilobular necrosis was observed. (C) In the 48 h group, moderate to marked centrilobular necrosis with inflammation were observed. (D) In the 72 h group, slight to moderate centrilobular necrosis was observed. H&E stain. 200× (A–D). Scale bars = 50 µm. |
 | Fig. 3Histopathological grading of the liver. In the 3 h and 6 h groups, liver histology appeared normal. Centrilobular necrosis with inflammation was first observed in the 24 h group. The lesion grade increased in the 48 h group and decreased in the 72 h group. Error bars indicate standard error of the mean (SEM). |
 | Fig. 4Time-course changes in miR-122 expression. In the APAP-treated groups, miR-122 expression was higher than in the vehicle group. The highest fold increase was observed in the 3 h group, whereas in the 72 group, the miR-122 level was similar to that in the vehicle group. Error bars indicate SEM. |
 | Fig. 5Time-course changes in serum chemistry. Some alanine transaminase (ALT) and, aspartate transaminase (AST) levels were minimally elevated in the APAP-treated group relative to the vehicle group (ALT at 48 h; AST at 6 h); however, the magnitude of the increase was small compared to the level in the vehicle group. Total bilirubin (TBIL) levels in the APAP-treated group were not significantly elevated, and TBIL levels were not toxicologically significant. Error bars indicate SEM. *p < 0.05, **p < 0.01. |
Acknowledgments
This work was supported by the Korean Health Technology R&D Project (grant Nos. HI06C0868 and HI10C2014), Ministry of Health & Welfare, Korea.
References
1. Amar PJ, Schiff ER. Acetaminophen safety and hepatotoxicity–where do we go from here? Expert Opin Drug Saf. 2007; 6:341–355.
3. Antoine DJ, Dear JW, Lewis PS, Platt V, Coyle J, Masson M, Thanacoody RH, Gray AJ, Webb DJ, Moggs JG, Bateman DN, Goldring CE, Park BK. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology. 2013; 58:777–787.

4. Assis DN, Navarro VJ. Human drug hepatotoxicity: a contemporary clinical perspective. Expert Opin Drug Metab Toxicol. 2009; 5:463–473.

5. Atkuri KR, Mantovani JJ, Herzenberg LA, Herzenberg LA. N-acetylcysteine–a safe antidote for cysteine/glutathione deficiency. Curr Opin Pharmacol. 2007; 7:355–359.
6. Bala S, Marcos M, Kodys K, Csak T, Catalano D, Mandrekar P, Szabo G. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor α (TNFα) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem. 2011; 286:1436–1444.

7. Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, Alao H, Kodys K, Szabo G. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. 2012; 56:1946–1957.

9. Castro RE, Ferreira DM, Zhang X, Borralho PM, Sarver AL, Zeng Y, Steer CJ, Kren BT, Rodrigues CMP. Identification of microRNAs during rat liver regeneration after partial hepatectomy and modulation by ursodeoxycholic acid. Am J Physiol Gastrointest Liver Physiol. 2010; 299:G887–G897.

10. Chalasani N, Fontana RJ, Bonkovsky HL, Watkins PB, Davern T, Serrano J, Yang H, Rochon J. Drug Induced Liver Injury Network (DILIN). Causes, clinical features, and outcomes from a prospective study of drug-induced liver injury in the United States. Gastroenterology. 2008; 135:1924–1934. 1934.e1–1934.e4.

11. Chong VH. Acetaminophen overdose and N-acetylcysteine therapy. Ann Acad Med Singapore. 2007; 36:704.
12. Craig DG, Bates CM, Davidson JS, Martin KG, Hayes PC, Simpson KJ. Staggered overdose pattern and delay to hospital presentation are associated with adverse outcomes following paracetamol-induced hepatotoxicity. Br J Clin Pharmacol. 2012; 73:285–294.

13. Finch ML, Marquardt JU, Yeoh GC, Callus BA. Regulation of microRNAs and their role in liver development, regeneration and disease. Int J Biochem Cell Biol. 2014; 54:288–303.

14. Imaeda AB, Watanabe A, Sohail MA, Mahmood S, Mohamadnejad M, Sutterwala FS, Flavell RA, Mehal WZ. Acetaminophen-induced hepatotoxicity in mice is dependent on Tlr9 and the Nalp3 inflammasome. J Clin Invest. 2009; 119:305–314.

15. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods. 2001; 25:402–408.

16. Marquez RT, Wendlandt E, Galle CS, Keck K, McCaffrey AP. MicroRNA-21 is upregulated during the proliferative phase of liver regeneration, targets Pellino-1, and inhibits NF-κB signaling. Am J Physiol Gastrointest Liver Physiol. 2010; 298:G535–G541.
17. O'Connell RM, Kahn D, Gibson WSJ, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010; 33:607–619.
18. Raschzok N, Sallmon H, Dame C, Sauer IM. Liver regeneration after partial hepatectomy: inconsistent results of expression screenings for human, mouse, and rat microRNAs. Am J Physiol Gastrointest Liver Physiol. 2012; 302:G470–G471.

19. Schug J, McKenna LB, Walton G, Hand N, Mukherjee S, Essuman K, Shi Z, Gao Y, Markley K, Nakagawa M, Kameswaran V, Vourekas A, Friedman JR, Kaestner KH, Greenbaum LE. Dynamic recruitment of microRNAs to their mRNA targets in the regenerating liver. BMC Genomics. 2013; 14:264.

20. Sgro C, Clinard F, Ouazir K, Chanay H, Allard C, Guilleminet C, Lenoir C, Lemoine A, Hillon P. Incidence of drug-induced hepatic injuries: a French population-based study. Hepatology. 2002; 36:451–455.

21. Starckx S, Batheja A, Verheyen GR, Jonghe SD, Steemans K, Van Dijck B, Singer M, Bogdan N, Snoeys J, Vinken P, Sasaki JC, Van Gompel J, Guzzie-Peck P, Lampo A, Lammens L. Evaluation of miR-122 and other biomarkers in distinct acute liver injury in rats. Toxicol Pathol. 2013; 41:795–804.

22. Starkey Lewis PJ, Dear J, Platt V, Simpson KJ, Craig DGN, Antoine DJ, French NS, Dhaun N, Webb DJ, Costello EM, Neoptolemos JP, Moggs J, Goldring CE, Park BK. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology. 2011; 54:1767–1776.

23. Tili E, Michaille JJ, Wernicke D, Alder H, Costinean S, Volinia S, Croce CM. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc Natl Acad Sci U S A. 2011; 108:4908–4913.

24. Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009; 106:4402–4407.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download