Introduction
Canine mammary gland tumors (CMGTs), which represent 50–70% of all tumors in sexually intact female dogs [18], affect middle-aged and older dogs and are most common in 6–14-year-old females [16]. Three main factors affect mammary tumor risk, age, hormonal exposure, and breed. In general, CMGTs are more common in smaller breeds [18].
The advantage of CMGTs as a model of human breast cancer arises from several similarities, including the relative age of onset, risk factors, incidence, histological and molecular features, biological behavior, metastatic patterns, and responses to therapy [11]. The same cytological criteria that are applied in human pathology have been reported to effectively diagnose CMGTs because they have similar cytological characteristics to human breast cancer [16]. Recent studies have examined microRNA (miRNA) expression in CMGTs [320]. MicroRNAs are small (21–25 nucleotides) non-coding RNAs. These molecules are believed to be involved in a wide range of developmental and cellular processes through base-pairing with complementary sites of target transcripts, either to repress translation or to trigger mRNA degradation [1]. Some miRNAs have been labeled "oncomirs" because of their associations with various cancers, and several studies have successfully used microarrays and quantitative real-time PCR to profile the miRNA expression patterns in several different cancers [2451219]. In humans, expressed miRNAs appear to correlate with specific breast cancer biopathologic features such as estrogen and progesterone receptor expression, tumor stage, vascular invasion, or proliferation index [4]. To the best of our knowledge, there have been few reports of miRNA expression in CMGTs. Therefore, the functions of miRNAs in CMGTs are not well understood. In the present study, we established a new cell line, the SNP cell line, from a CMGT and examined the expression levels of miRNAs in the cultured cells.
Materials and Methods
Ethics statement
The use of the animals and procedures employed were approved by the Animal Research Committee of Kitayama Labes (project No. GL50039). The study met the guidelines of the Institute of Laboratory Animal Resources for the use of experimental animals. All efforts were made to minimize suffering.
Case history
A 10-year-old mixed breed dog presented at the Veterinary Teaching Hospital of Tottori University with a mass in the left mammary gland. The tumor size was approximately 4 × 6 × 14 cm. Chest radiographs showed the presence of pleural effusion. The tumor and axillary lymph node were removed surgically and fixed for histopathology. The pleural effusion was removed by thoracentesis. Tumor cells were observed in the pleural effusion and used for primary culture.
Histology
The original mammary gland tumor tissue and tumors established in subcutaneous tissue of NOD.CB17-Prkdcscid/J mice were fixed in 10% buffered formaldehyde, embedded in paraffin, and sectioned and stained with hematoxylin and eosin (H&E).
Establishment of the cell line and doubling time
The tumor cells were prepared under aseptic conditions for cell culture. The cells were cultured in 35-mm diameter petri dishes (Nunc, Denmark) in RPMI 1640 medium (Invitrogen, USA) supplemented with 10% heat-inactivated fetal bovine serum (Nichirei Biosciences, Japan) and PSN (5 mg/mL penicillin, 5 mg/mL streptomycin, and 10 mg/mL neomycin) solution (Invitrogen), then incubated in 5% CO2 at 37℃. The cells were washed with phosphate-buffered saline for subculture. Next, the cells were harvested from near-confluent cultures by brief exposure to a solution containing 0.25% trypsin and 1 mmol/L tetrasodium ethylenediaminetetraacetic acid solution with phenol red (Invitrogen). Trypsinization was stopped using RPMI 1640 containing 10% fetal bovine serum. Trypsinized cells were transferred to a new petri dish.
For clonal experiments, cell suspensions were serially diluted and plated onto 96-well plates. Wells containing one single cell after microscopic confirmation were marked. A colony was then sub-cultured from each marked well into a 12-well plate. Cloning was repeated two times. Cell lines were maintained in continuous culture over 60 passages, and the established cell line was designated as SNP cells.
SNP cells at passage 55 were used for the growth assay. Briefly, cells were incubated in 35 mm diameter petri dishes at 5 × 104 cells/mL in 2 mL of cultivation medium for 24 h. Following incubation, cells were harvested and prepared for daily cell counts, which were conducted using a Muse Count & Viability Kit (EMD Millipore, USA) according to the instructions supplied by the manufacturer. Cells were co-incubated with Muse Count & Viability Reagent at room temperature for 5 minutes in the dark. Single-cell suspensions were loaded onto the instrument, and viable cells were counted. The doubling time was calculated using the following equation:
where N1 is the number of seeded cells and N2 is the number of counted cells at time h.
To assess their morphological features, SNP cells were incubated in 35 mm diameter petri dishes incubated in 5% CO2 at 37℃. The cells were visualized under an OLYMPUS Fluoroview 1000 Confocal Laser Microscope (Olympus, Japan).
Tumorigenicity
Two NOD.CB17-Prkdcscid/J mice (female, 6 weeks old) were purchased from Charles River Laboratories International (Japan). The animals were maintained under conventional conditions and provided with a standard pellet diet and water ad libitum. SNP cells at passage 48 were inoculated subcutaneously into the ventral flank of each mouse at a concentration of 1.0 × 107 cells/0.1 mL using a 26-gauge syringe. The mice were sacrificed by cervical dislocation 21 days after inoculation, after which the tumors were harvested. The specimens were fixed in 4% buffered formalin, embedded in paraffin, and sectioned and stained with H&E.
Immunohistochemistry
The sections from the tumors derived from mice were incubated with anti-vimentin monoclonal antibody (clone V9; YLEM, Italy) followed by the peroxidase-labeled secondary antibody (Simple stain MAX PO; Nichirei Biosciences, Japan). The signals were visualized by the 3,3'-diaminobenzidine and the nuclei were counterstained using Mayer's hematoxylin.
miRNA arrays
SNP cells or normal mammary gland tissue were freshly frozen within 30 minutes of harvesting or surgical excision and stored at -80℃ until further analysis using an Affymetrix GeneChip miRNA array. An Agilent 2100 Bioanalyzer (Agilent Technologies, USA) was used to assess the quality of total RNA (1,000 ng). Biotin-labeled samples were prepared with a Flash Tag Biotin HSR RNA Labeling Kit (Affymetrix, USA) according to the manufacturer's protocol, and 21.5 µL of biotin-labeled sample was combined with 110.5 µL of Hybridization Master Mix. The array was incubated at 48℃ and 0.3 × g for 18 h in a GeneChip Hybridization Oven 645 (Affymetrix) according to the manufacturer's protocol. A GeneChip Fluidics Station 450 (Affymetrix) was used to wash the array according to the manufacturer's protocol, after which it was scanned with a GeneChip Scanner 3000 7G (Affymetrix) following the protocol in the Affymetrix GeneChip Command Console User Manual.
Results
Histology
Epithelial neoplastic cells with nuclei proliferated in a tubulopapillary or trabecular pattern in the expanded milk ducts. The cells were characterized by irregularly sized nuclei, clarification or increased size of nucleoli, multilayered arrangement, and blood or lymphatic vessel invasion of neoplastic cells. In addition, extensive necrosis was scattered throughout the neoplastic tissue (Fig. 1). Based on these histopathological findings, the tumor was diagnosed as a CMGT.
Characterization of the established cell line and doubling time
The doubling time at passage 57 was 26.2 h, and the cell line was cultured for over 60 passages. SNP cells exhibited round to ovoid prominent nuclei, a round to short spindle shape, and ample cytoplasm.
Tumorigenicity
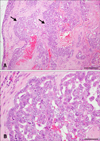
A tumor formed in each mouse at the site of subcutaneous SNP cell injection. Histologically, the circumscribed neoplastic nodules were located in the dermis and subcutaneous tissue. The tumor was composed of luminal epithelium-like cells, and the neoplastic cells were arranged with a tubulopapillary architecture. The nuclei were round to ovoid and normochromatic to hyperchromatic with prominent nucleoli. Cell borders were distinct. Mitotic figures and pyknotic nuclei were observed for a large proportion of the cells (Fig. 2). Histopathologically, the morphological features of SNP cells were similar to those seen in the original CMGT cells. Some of the neoplastic cells showed positive signals for vimentin (panel C in Fig. 2). These findings were marked in the marginal region of the mass, as well as in the neoplastic cells within the vessels.
miRNA arrays
All samples were analyzed using the Agilent 2100 Bioanalyzer, and the 18S and 28S ribosomal RNA peaks were identified. The RNA integrity of SNP cells and normal mammary gland tissue were 9.6 and 7.2, respectively.
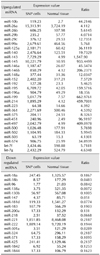
Comparison of samples from SNP cells and normal mammary gland tissue revealed 291 different miRNAs. Table 1 lists 48 miRNAs that showed large (> 4-fold) increases (30 miRNAs) or decreases (18 miRNAs) in expression in SNP cells relative to normal mammary gland tissue. Notably, the expression of miRNA-143 and miRNA-138a showed the largest increase and decrease, respectively.
Discussion
In this study, we successfully established a novel SNP cell line that retained the morphological characteristics of canine tumor cells. Previous work has suggested that the vast similarities in both genetics and physiology and the considerable overlap in treatment regimens make canines an ideal model for many human diseases, including malignant breast cancer [3]. Therefore, SNP cells might serve as a model for future genetic analysis and treatment of humans with malignant breast tumors.
Detailed understanding of the dysregulation of miRNAs in cancer might be useful for diagnosis, prediction of prognosis, and therapy. In this study, we also analyzed SNP cells and normal mammary gland tissue to identify miRNAs with altered expression in SNP cells and found 30 that increased by > 4-fold and 18 that decreased by > 4-fold.
In this study, the expression of miRNA-143 showed the greatest increase in SNP cells, with expression 1,547.9-fold above that of normal mammary gland tissue. miRNA-143 was previously shown to have significantly increased expression in mammary tumor tissue (non metastasizing carcinoma) relative to normal mammary tissue, with an expression difference of 2.70-fold [20]. These results suggested that miRNA-143 might be an oncogenic miRNA. Additionally, miRNA-10b expression was increased 44.2-fold between the SNP cell line and normal mammary gland tissue. MiRNA-10b was identified as a tumor suppressor that prevents human breast cancer development, as well as an oncogene that initiates breast cancer invasion and metastasis [10], and its expression is increased in neoplasms [9]. Overexpression of miRNA-10b promotes cell migration and invasion in vitro. The transcription factor Twist likely induces expression of miRNA-10b, which can then target the transcription factor homeobox D10 and inhibit its translation, resulting in induction of the pro-metastatic gene product, Ras homologue gene family member C [17]. Furthermore, the expression of miRNA-29b was increased 4.0714-fold in the SNP cell line compared to normal mammary gland tissue. A previous report demonstrated that miRNA-29b was significantly up-regulated in cancerous CMGT samples relative to normal tissue [3]. Although there is little data available regarding miRNAs in CMGTs, we suggest that up-regulation of miRNA-10b, miRNA-29b, and miRNA-143 might be associated with CMGT based on our data reported herein and previous observations of expression changes in these miRNAs in tumor tissues.
Several other miRNAs also showed increased expression in SNP cells. The expression of miRNA-145 increased 933.4-fold over that of normal mammary gland tissue expression. However, miRNA-145 was reportedly progressively down-regulated in cancer cells with a high proliferation index, canine malignant melanoma tissues, and canine and human melanoma cell lines [15]. Similarly, let-7f and let-7g increased 5.7-fold and 4.6-fold, respectively, in SNP cells compared to normal mammary gland tissue. The expression of various let-7 miRNAs was previously observed to be down-regulated in breast cancer samples with either lymph node metastasis or a high proliferation index, suggesting that reduced let-7 expression could be associated with poor prognosis [4]. Therefore, we propose that miRNA-145, let-7f, and let-7g might not play a significant role as suppressors of SNP cells.
The greatest decrease in expression was observed for miRNA-138a, which showed a 0.007-fold difference between SNP cells and normal mammary gland tissue. Previously, miRNA-138a was shown to suppress the epithelial to mesenchymal transition (EMT) in squamous carcinoma cell lines and regulate cell migration and invasion [8]. In this study, some SNP cells showed positive signals for vimentin. In recent years, vimetin has gained importance as a marker of EMT [21]. Therefore, these findings indicate that SNP cells undergo the EMT process. MiRNA-138a has also been implicated as a tumor suppressor, potentially through targeting neutrophil gelatinase-associated lipocalcin or hTERT [6714]. Thus, down-regulation of miRNA-138a might be associated with CMGT.
The expression of miRNA-221 and -222 differed by 0.2187- and 0.1944-fold, respectively, between SNP cells and normal mammary gland tissue. The miRNA-221/222 cluster regulates the cell cycle, cell growth, and EMT in breast cancer. It has been reported that the expression of miRNA-221/222 was significantly (2-fold) elevated in HER2/neu-positive primary human breast cancer tissues known to be resistant to endocrine therapy when compared with HER2/neu-negative tissue samples [13]. Hence, we suggest that miRNA-221/222 might not play a significant role in the tumorigenesis of SNP cells.
In conclusion, our newly established SNP cells from a CMGT are characterized by proliferation in a tubulopapillary pattern and vimentin positive. Moreover, our results show that the expression values of miRNA-143 and miRNA-138a have the greatest increase and decrease, respectively, in SNP cells. Further research using SNP cells may allow advances in the diagnosis and treatment of CMGTs by providing gene technology using miRNAs. Moreover, because CMGTs have been suggested as a model for human breast cancer and EMT process, SNP cells might serve as a model for future genetic analysis and clinical treatment of human breast tumors.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


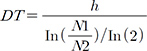
 XML Download
XML Download