Abstract
Edwardsiella (E.) ictaluri is a major bacterial pathogen that affects commercially farmed striped catfish (Pangasius hypothalamus) in Vietnam. In a previous study, 19 strains of E. ictaluri collected from striped catfish were biochemically identified with an API-20E system. Here, the same 19 strains were used to assess the ability of matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS; applied using a MALDI Biotyper) to conduct rapid, easy and accurate identification of E. ictaluri. MALDI-TOF MS could directly detect the specific peptide patterns of cultured E. ictaluri colonies with high (> 2.0, indicating species-level identification) scores. MALDI Biotyper 3.0 software revealed that all of the strains examined in this study possessed highly similar peptide peak patterns. In addition, electrophoresis (SDS-PAGE) and subsequent immuno-blotting using a specific chicken antibody (IgY) against E. ictaluri revealed that the isolates had highly similar protein profiles and antigenic banding profiles. The results of this study suggest that E. ictaluri isolated from striped catfish in Vietnam have homologous protein compositions. This is important, because it indicates that MALDI-TOF MS analysis could potentially outperform the conventional methods of identifying E. ictaluri.
The striped catfish (Pangasius hypophthalmus) is one of the most important cultured fish species in Vietnam [4]. Emerging evidence suggests that bacillary necrosis of Pangasianodon (BNP), which is caused by Edwardsiella (E.) ictaluri, is responsible for severe economic losses, especially in fingerlings and juvenile stripped catfish [34]. Intensive rearing of striped catfish in the Mekong Delta might have led to increased outbreaks of BNP observed in recent decades. The major pathological signs of E. ictaluri infection include multifocal irregular white nodules of varying sizes on the liver, spleen and kidney of the striped catfish. Moreover, the disease is characterized by a high mortality rate [7].
E. ictaluri is also a well-known bacterial pathogen responsible for enteric septicemia of channel catfish (Ictalurus punctatus) in the United States [89]. A previous study compared E. ictaluri isolates from striped catfish of Vietnam with those from channel catfish of the United States [16]. The study revealed that the isolates possessed biochemically similar characteristics, as they were both fermentative in glucose oxidation/fermentation (O/F) medium, catalase positive and oxidase negative upon analysis using the API 20E system (Vitek 2; bioMérieux, France), and showed homologous fingerprinting patterns in genetic analyses using repetitive sequence-based polymerase chain reaction (rep-PCR) [16].
In an effort to control and prevent outbreaks of BNP, researchers have attempted to develop rapid techniques for identifying E. ictaluri infection. PCR-based methods have been designed to rapidly detect pathogenic bacteria in fish [17], and evidence suggests that multiplex PCR could be useful for simultaneous detection of major bacterial fish pathogens, such as Flavobacterium columnare, E. ictaluri, and Aeromonas hydrophila [14]. In addition, an ELISA-based technique was developed to detect and quantify E. ictaluri in sub-clinically infected channel catfish, and the results were found to be comparable to those obtained from conventional bacterial culturing methods [5].
Recently, matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) was introduced as a method for rapidly identifying bacterial strains isolated from clinical samples [2]. This method uses various algorithms to compare the spectral fingerprint of an unknown isolate with a database of reference spectra, regardless of small between-colony and culture-condition-related differences [11]. Here, we assessed the use of a MALDI-TOF MS system (MALDI Biotyper; Bruker Daltonik, Germany) as a rapid, reliable and accurate means for identifying E. ictaluri strains isolated from striped catfish of Vietnam.
Nineteen E. ictaluri strains isolated from striped catfish in Vietnam's Mekong Delta and reference strain ATCC33202 isolated from channel catfish were examined in this study [16]. All strains were cultured in brain heart infusion broth (Becton, Dickinson and Company, USA) or trypticase soy agar (Becton, Dickinson and Company) supplemented with 5% sheep blood (Thermo Scientific, USA) for 48 h at 28℃. Biochemical methods were used to confirm the identification of E. ictaluri, including Gram staining, cytochrome oxidase assays, catalase assays, glucose fermentation assays, glucose motility dips and analysis on an API 20E system (bioMérieux), as previously described [16].
E. ictaluri colonies were suspended in 300 µL of distilled water and mixed with 900 µL of absolute ethanol. After vortexing, the samples were centrifuged at 16,000 × g for 2 min, and the supernatant was decanted. The pellets were then resuspended in 500 µL of distilled water and centrifuged at 16,000 × g for 2 min. The supernatant was removed, mixed with 50 µL of absolute acetonitrile and 50 µL of 70% formic acid, and vortexed for 2 min. The sample was then centrifuged as described above, and 1 µL of the supernatant was targeted onto an MSP (main spectra library) 96-target polished steel plate (Bruker Daltonik). Next, the sample was dried, and 1 µL of α-cyano-4-hydroxycinnamic acid matrix solution (saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile and 2.5% trifluoroacetic acid) was added to the sample and allowed to crystalize.
Mass spectra were acquired using a microflex LT mass spectrometer (Bruker Daltonik) managed by the flexControl software (ver. 3.0; Bruker Daltonik). Positive ions were extruded with an accelerating voltage of 20 kV, and spectra were analyzed within mass/charge (m/z) ratios of 2,000 to 20,000 in positive linear mode. Each spectrum was calibrated with respect to a bacterial test standard (255343; Bruker Daltonik). The acquired data were measured and analyzed with the MALDI Biotyper 3.0 system, using the Bruker database. A logarithmic score of 0 to 3.0 was assigned according to spectral peak matching patterns and the acquired score means as follows: a score of 0 to 1.699 indicated no reliable identification; a score of 1.700 to 1.999 indicated probable genus-level identification; a score of 2.000 to 2.299 indicated secure genus-level identification and probable species-level identification; and a score of 2.300 to 3.000 indicated highly probable species-level identification.
Edwardsiella ictaluri 258 (a representative strain) isolated from striped catfish in Vietnam was grown in tryptic soy broth (TSB) for 48 h, inactivated with 1% formalin for 24 h at 4℃, centrifuged at 7,500 × g for 20 min, washed three times with sterile phosphate-buffered saline (PBS) [3 mM KCl, 137 mM NaCl, 1.5 mM KH2PO4 and 8 mM Na2HPO4 (pH 7.2)] and resuspended in PBS. Chickens were the immunized as previously described, with some modifications[10]. Initially, 1.0 × 108 colony-forming units (CFU)/mL of formalin-killed 258 strain emulsified with an equal volume of Freund's Complete Adjuvant (Sigma, USA) were immunized. The chickens were then immunized with 1.0 × 108 CFU/mL of formalin-killed 258 strain emulsified with Freund's incomplete adjuvant three times at intervals of 2 weeks. One week after the last immunization, chicken eggs were collected, and chicken IgY was purified from the eggs using an EGGstract IgY Purification System (Promega, USA) according to the manufacturer's instructions. The purified IgY pellet was resuspended in PBS and stored at −20℃ until use.
Each of the studied E. ictaluri strains was grown in TSB for 48 h, inactivated with 1% formalin for 24 h at 4℃, centrifuged at 7,500 × g for 20 min, washed three times with PBS and resuspended to 1.0 × 109 CFU/mL in PBS. The above-described chicken anti-E. ictaluri IgY was diluted serially two-fold in PBS, from 1 : 2 to 1 : 16,384, and 50 µL of each dilution was transferred to wells of a U-bottomed 96-well microplate. An equal volume of formalin-killed E. ictaluri was added, and the plate was incubated overnight at 4℃. The last dilution that induced agglutination was recorded. Each individual strain was tested in triplicate.
SDS-PAGE was performed on 12.5% (w/v) separating gels, according to the standard method [12]. Bacterial isolates were cultured in TSB at 28℃ for 48 h, centrifuged at 2000 × g for 30 min, washed three times with PBS, resuspended in 200 µL of PBS, and mixed with 50 µL of 5 × sample buffer [5 : 1; 60 mM Tris-HCl, 25% (v/v) glycerol, 2% (w/v) SDS, 14.4 mM 2-mercaptoethanol, and 0.1% (w/v) bromophenol blue]. The samples were then sonicated 10 times (5.5 W, 10 sec intervals; XL-2020; Misonix, USA), boiled for 10 min, cooled on ice, and centrifuged at 16,000 × g for 20 min at 4℃. The supernatants were collected and electrophoresed on SDS gels at 50 V for the first 15 min, then at 100 V until the leading line reached the bottom of the gel. The gels were subsequently stained with Coomassie Brilliant Blue R-250.
The resolved proteins were blotted to polyvinylidenedifluoride membranes at 70 V for 70 min. Next, the membranes were soaked in 100% methanol for 20 sec, dried at room temperature (RT), blocked with 5% (w/v) skim milk in PBS-T [3 mM KCl, 137 mM NaCl, 1.5 mM KH2PO4, 8 mM Na2HPO4 (pH 7.2), and 0.05% (v/v) Tween-20] for 60 min at RT, washed three times with PBS-T, and incubated with anti-E. ictaluri chicken IgY (1 : 50,000) for 60 min at RT. After three washes with PBS-T, the membranes were incubated with rabbit anti-chicken IgG-HRP (1 : 20,000; Cappel Biologicals, USA) for 90 min at RT. After three washes with PBS-T (15 min each), the membrane was developed with an enhanced chemiluminescence kit (GE Healthcare Life Science, USA) and used to expose an X-ray film.
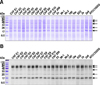
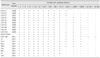
Nineteen previously characterized E. ictaluri strains and reference strain ATCC33202 were analyzed by MALDI-TOF MS, and the obtained spectra were compared with those in the Bruker database. All of the examined strains were identified as E. ictaluri at the species level. Specifically, 65% (13/20) were identified with log scores > 2.3, while 35% (7/20) had log scores between 2.0 and 2.3. No strain showed a log score < 2.0 (Table 1). It took less than 60 min from initial sample preparation to obtain the identification results. Importantly, while the mass intensities were diverse, most of the peak patterns were quite similar among the striped catfish isolates and ATCC33202 (e.g., peaks at 2658, 5321, 6259, 9281, 9465 and 11,542 Da) (Fig. 1).
Specific chicken IgY was produced using E. ictaluri strain 258 and used as a recognition agent for serological agglutination tests. The agglutination assay exhibited a sensitivity of 79% for identifying E. ictaluri isolates at a 1 : 1,024 sample dilution, and 100% positive reactions were achieved with a 1 : 256 dilution (Table 2). Strain ATCC33202 was also agglutinated with chicken IgY (1 : 4,096 sample dilution).
SDS-PAGE was used to evaluate the between-strain diversity of the protein profiles. Coomassie Brilliant Blue R-250 staining demonstrated that the resolved protein bands were quite similar in molecular weight at 17–62 kDa, except for 23 kDa, for which some isolates possessed high or low intensity. There were no distinguishable bands < 17 kDa (panel A in Fig. 2).
Subsequent immunoblot assays were conducted with the chicken IgY, which successfully recognized the resolved E. ictaluri proteins. The antigenic profiles of E. ictaluri lysates were very similar to one another in the molecular masses of their antigenic bands (e.g., at 62, 55, 40, 25 and 17 kDa). There was no significant between-lysate differences in band intensity (panel B in Fig. 2).
E. ictaluri infection in striped catfish is a major concern in the fresh-water culture industry of Vietnam [4]. To remedy the severe economic losses sustained by this important fish stock, effective diagnostic methods for E. ictaluri infection have been developed based on genomic and proteomic techniques that specifically detect genetic loci and antigens of E. ictaluri [1417]. Despite their specificity, these methods are time consuming. Recent publications on various pathogenic bacteria have suggested that MALDI-TOF MS could be a high-throughput screening method that could allow simultaneous identification of a number of bacteria within one hour [211]. Here, all of the E. ictaluri isolates from striped catfish were successfully identified with a high degree of confidence using a MALDI Biotyper. Thus, the present study indicates that MALDI-TOF MS could be a versatile technology for detecting E. ictaluri based on the conserved characteristics of its proteomic composition.
In general, two types of sample preparation methods are recommended when using a MALDI Biotyper to acquire bacteria-specific mass peak patterns [26]. The first is the direct sample preparation method, in which colonies are obtained from fresh cultured agar plates and applied directly onto the target plate, while the second is the extraction method, which was applied in this study. Both procedures are quick and simple, and were thus carefully compared for their ability to obtain high-quality and reproducible results. Our results revealed that the direct sample preparation method produced lower scores than the extraction method (data not shown), with 20% of the isolates (4/20) yielding scores < 2.0, meaning that they could only be identified to the genus level. A previous study using bacteria routinely isolated from a hospital setting demonstrated that the extraction method yielded valid identification scores 25% more often than the direct sample preparation method [2]. Thus, it appears that, despite the simplicity of the direct preparation method, the extraction method is preferable for the generation of higher scores and prevention of false negative identifications.
Notably, increasing evidence suggests that MALDI-TOF MS does not guarantee accurate species-level identification for some bacteria. Acinetobacter nosocomialis, which is closely related to members of the Acinetobacter baumannii group, was not identified using the MALDI Biotyper approach because the Bruker database system did not contain the necessary spectral information [6]. Furthermore, pathogenic Mycobacterium (M.) marinum strains isolated from marine fish were not successfully identified, even though the Bruker database included reference information regarding M. marinum [11]. In the present study, the examined strains were successfully identified as E. ictaluri, with high scores (2.04–2.429) obtained against the Bruker database. Peak analysis of spectra obtained from the 19 tested strains of E. ictaluri and ATCC33202 revealed high similarities among many of their peaks, such as those found at 2658, 5321, 6259, 9281, 9465 and 11,542 Da. This conserved species-specific protein signature may contribute to the ability of MALDI-TOF MS to successfully identify E. ictaluri.
The results of our SDS-PAGE analysis and Western blot assays using specific chicken IgY were highly consistent with the MALDI-TOF MS-based findings of spectral consistency among E. ictaluri isolates. SDS-PAGE indicated that the 20 strains of E. ictaluri (including ATCC33202) comprised a homologous protein group that shared at least 10 similar protein bands with molecular weights of 17–62 kDa. The Western blot assay revealed consistent bands at approximately 62, 55, 40, 25 and 17 kDa in all 20 E. ictaluri strains, as recognized by chicken IgY. Moreover, the IgY was agglutinated by all strains of E. ictaluri at a 1 : 256 dilution. Our results are consistent with early reports indicating that the various isolates of E. ictaluri yielded homogenous results for proteomic and serological analyses, such as SDS-PAGE, Western blotting and agglutination assays [11315]. These observations provide further evidence indicating that the proteins comprising E. ictaluri are homogenous, making this pathogen a good candidate for MALDI-TOF MS-based identification.
In conclusion, we demonstrated that MADLI-TOF MS could be a valuable tool for identifying E. ictaluri from field samples. The 20 strains tested herein (19 strains isolated from striped catfish, plus the reference strain) all exhibited highly homogenous proteomic compositions, as assessed by MADLI-TOF MSbased peak analysis and the examination of protein band patterns obtained upon SDS-PAGE and Western blot analysis. The MALDI-TOF MS-based method is simpler and easier than currently utilized genomic approaches (e.g., PCR and sequencing), and might therefore facilitate laboratory diagnosis of E. ictaluri infections in the aquaculture industry.
Figures and Tables
Fig. 1
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) spectral profiles of three representative Edwardsiella (E.) ictaluri strains, as generated using the flex Analysis software and a MALDI Biotyper system. The relative intensities of the ions are shown on the Y-axis, and the ion masses (in Daltons) are shown on the X-axis. The m/z value gives the mass-to-charge ratio. Peptide peaks common to all strains are indicated with arrow heads above the panel, and the peaks with black lines in the panel represent different peaks between the four strains. a.u., arbitrary units.

Fig. 2
Electrophoresis (SDS-PAGE) (A) and Western blot (B) analysis of whole cell lysates from E. ictaluri isolated from striped catfish. Black arrows indicate consistent bands of whole cell lysates.
Acknowledgments
This work was made possible by a Korea Research Foundation grant funded by the Ministry of Science, ICT and Future Planning of Korea (NRF-2013R1A1A1059608), and by the Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA) and Korea Forest Service (KFS) (213004043SB310 and 213004-04-3-SB320).
References
1. Bertolini JM, Cipriano RC, Pyle SW, McLaughlin JJ. Serological investigation of the fish pathogen Edwardsiella ictaluri, cause of enteric septicemia of catfish. J Wildl Dis. 1990; 26:246–252.

2. Bizzini A, Durussel C, Bille J, Greub G, Prod'hom G. Performance of matrix-assisted laser desorption ionizationtime of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol. 2010; 48:1549–1554.

3. Crumlish M, Thanh PC, Koesling J, Tung VT, Gravningen K. Experimental challenge studies in Vietnamese catfish, Pangasianodon hypophthalmus (Sauvage), exposed to Edwardsiella ictaluri and Aeromonas hydrophila. J Fish Dis. 2010; 33:717–722.

4. Dung TT, Haesebrouck F, Tuan NA, Sorgeloos P, Baele M, Decostere A. Antimicrobial susceptibility pattern of Edwardsiella ictaluri isolates from natural outbreaks of bacillary necrosis of Pangasianodon hypophthalmus in Vietnam. Microb Drug Resist. 2008; 14:311–316.

5. Earlix D, Plumb JA, Rogers W. Isolation of Edwardsiella ictaluri from channel catfish by tissue homogenization, filtration and enzyme linked immunosorbent assay. Dis Aquat Organ. 1996; 27:19–24.

6. Espinal P, Seifert H, Dijkshoorn L, Vila J, Roca I. Rapid and accurate identification of genomic species from the Acinetobacter baumannii (Ab) group by MALDI-TOF MS. Clin Microbiol Infect. 2012; 18:1097–1103.

7. Ferguson HW, Turnbull JF, Shinn A, Thompson K, Dung TT, Crumlish M. Bacillary necrosis in farmed Pangasius hypophthalmus (Sauvage) from the Mekong Delta, Vietnam. J Fish Dis. 2001; 24:509–513.

8. Hawke JP. A bacterium associated with disease of pond cultured channel catfish, Ictalurus punctatus. Can J Fish Aquat Sci. 1979; 36:1508–1512.

9. Hawke JP, McWhorter AC, Steigerwalt AG, Brenner DJ. Edwardsiella ictaluri sp. nov., the causative agent of enteric septicemia of catfish. Int J Syst Bacteriol. 1981; 31:396–400.

10. Kang SH, Shin G, Shin Y, Palaksha KJ, Kim Y, Yang H, Lee E, Lee E, Huh N, Ju OM, Jung T. Experimental evaluation of pathogenicity of Lactococcus garvieae in black rockfish (Sebastes schlegeli). J Vet Sci. 2004; 5:387–390.

11. Kurokawa S, Kabayama J, Fukuyasu T, Hwang SD, Park CI, Park SB, del Castillo CS, Hikima J, Jung TS, Kondo H, Hirono I, Takeyama H, Aoki T. Bacterial classification of fish-pathogenic Mycobacterium species by multigene phylogenetic analyses and MALDI Biotyper identification system. Mar Biotechnol (NY). 2013; 15:340–348.

12. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970; 227:680–685.

13. Lobb CJ, Ghaffari SH, Hayman JR, Thompson DT. Plasmid and serological differences between Edwardsiella ictaluri strains. Appl Environ Microbiol. 1993; 59:2830–2836.

14. Panangala VS, Shoemaker CA, Van Santen VL, Dybvig K, Klesius PH. Multiplex-PCR for simultaneous detection of 3 bacterial fish pathogens, Flavobacterium columnare, Edwardsiella ictaluri, and Aeromonas hydrophila. Dis Aquat Organ. 2007; 74:199–208.

15. Plumb JA, Klesius P. An assessment of the antigenic homogeneity of Edwardsiella ictaluri using monoclonal antibody. J Fish Dis. 1988; 11:499–509.

16. Rogge ML, Dubytska L, Jung TS, Wiles J, Elkamel AA, Rennhoff A, Oanh DTH, Thune RL. Comparison of Vietnamese and US isolates of Edwardsiella ictaluri. Dis Aquat Organ. 2013; 106:17–29.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download