Abstract
Animal models, particularly pigs, have come to play an important role in translational biomedical research. There have been many pig models with genetically modifications via somatic cell nuclear transfer (SCNT). However, because most transgenic pigs have been produced by random integration to date, the necessity for more exact gene-mutated models using recombinase based conditional gene expression like mice has been raised. Currently, advanced genome-editing technologies enable us to generate specific gene-deleted and -inserted pig models. In the future, the development of pig models with gene editing technologies could be a valuable resource for biomedical research.
The use of animal models has been a valuable tools in both basic science and in vivo studies. Initially for finding animal models, a natural mutated individual was selected and the inbreeding was used for increasing the population. Another way is to produce animal models by chemicals or drug treatment. It has been very limited as to secure a sufficient number or consistent phenotype of the models. Thus, the best approach for obtaining animal models is use of genetically modified animals. Along with the isolation of germ-line transmitted murine embryonic stem cell lines [6], animal models, particularly in mice, can now be rapidly generated. As a result, thousands of mouse models have been developed for biomedical research.
However, the use of mice has many disadvantages, including their small size, short-life span and multiple differences from human metabolism. These shortcomings have brought about a need for large animal models. In particular, pig models have been used in translational biomedical research because they have many anatomical and physiological similarities with humans [10]. For example, several pig models have been actively developed, investigated, and used for clinical research in areas such as organ transplantation in the xenotransplantation field [26]. They have also been utilized in studies involving cancer [7], neuronal [1634] and metabolic models.
Unlike mice models, there is still remained to improve in the development of multiple genetically modified porcine models [10]. The first transgenic pigs were generated by DNA microinjection [11]. However, this technique has low efficiency and various gene expressional levels (mosaicism) [5], which has led to somatic cell nuclear transfer (SCNT) being the preferred for developing transgenic pigs [25]. To produce transgenic pigs via SCNT, donor cells are transfected with exogenous DNA. In an initial SCNT study, fluorescent expressing piglets were generated through transfected donor cells [19]. Since then, various consistent trials for DNA engineering, transfection, and cell cultures have enabled us to produce multiple genes expressing piglets, even knockout (KO) pigs, via homologous recombination (HR). Even though these process is still inefficient, advances in SCNT based on improving in vitro maturation, activation conditions, and culture have accelerated the development of pig models for biomedical research. Recently, conditional transgenic pigs have been created by tetracycline-dependent gene expression and genome-editing technologies including DNA endonucleases (ZFN and TALEN) [17], in which every gene that can be edited theoretically has been inserted into pig genome. The purpose of this review is to examine the current state of transgenesis and genome-editing technologies in producing pig models for biomedical research.
The first transgenic pig using microinjection has been generated [11]. As described above, most transgenic pigs have been produced by SCNT with mutated cell lines. Recently, KO and Knockin (KI) pig models have also been generated via homologous recombination and genome-editing technologies. Recent scientific developments have led to the use of pig models in several specific fields, as summarized in Table 1.
Simply constitutive or tissue-specific promoter dependent overexpression and conditional gene-regulation systems including recombinase- dependent gene expression are necessary to produce better transgenic pig models.
Overexpression: For overexpression, constitutive promoters, primarily CMV, EF1α and CAG, were utilized for expression vector construction with the target gene and as selection markers. Early studies employed transgenic pig models based on simple transgene overexpression using constitutive promoters. However, the use of this approach has been reduced because constitutive expression may cause unexpected damage to transgenic animals. Therefore, tissue-specific promoters are used as an alternative. Initially, using a tissue-specific promoters from mice or human, transgenic pig models were generated [24]. Subsequently, a specific tissue promoter for transgenic pigs was developed and used [15]. In the future, a greater diversity of porcine-specific tissue promoters should be developed as higher genetic models.
Conditional gene expression: Ubiquitous expression in transgenic animals may be lethal in early embryonic development or not be different from genetic expression pattern because some genes will be expressed in the adult stage or under specific conditions. Because of these reasons, an increasing number of involving studies on conditional gene expressions such as Cre-loxP and Tet-on/off have been widely applied to mimic the disease or gene function in mice. In contrast, conditional gene expression models in pigs remain limited. This review considers the following gene-regulation systems utilizing experimental data: Cre-loxP, Dre-rox, PhiC31 and Tet-on/off systems.
Cre-loxP. Cre-loxP is the most widely employed system for generating conditional gene expression. Cyclic recombinase (Cre) recognizes specific sequences, named loxP, that are composed of 34 bps including an 8 bps asymmetric core region enclosed by two 13 bps inverted repeat regions and cause excision, insertion, inversion and translocation [323]. Due to this genome conditional engineering, Cre-loxP has primarily been applied in generating conditional transgenic mice [4]. In contrast, its use in producing large animals has been very limited. However, as the importance of pig models in biomedical research has increased, so has interest in producing conditional pig models using Cre-loxP.
Recently, transgenic pig research has been carried out using the Cre-loxP system [8202122]. In those studies, gene excision and insertion were successfully completed using Cre recombinase. Additionally, our study confirmed the viability of using Cre recombinase to execute gene cassette exchanges (Fig. 1). If transgenic pigs can be generated via SCNT using cassette exchangeable donor cells, then various genetic functions with no change in expression level can be analyzed after gene exchange.
Dre-rox. Recently, another site-specific recombinase, Dre, was identified in P1-like phages. Like Cre, Dre recombinase recognizes the specific sequence, rox, and causes excision of the flanked gene. Although Dre recombinase has a similar structure to Cre, it does not recognize loxP sequences, indicating that there is no crossover-recombination between Cre-rox and Dre-loxP [1]. In a study, the Cre and Dre recombination were used to produce a double conditional gene expression mouse model for retinal ganglion cell labeling [30]. However, Dre-rox recombination in pigs has not yet been investigated. As a preliminary study, our research group used porcine fibroblasts and embryos to excise the flanked fluorescence gene, rox, utilizing Dre recombinase (Fig. 2). Dre-rox can be another valuable tool conditional gene regulation in pigs.
PhiC31 recombinase. Unlike Cre- and Dre-recombinase with various genome engineering functions, PhiC31 recombinase can integrate the target gene into a site-specific sequence region, attP. PhiC31 protein recombines attB and attP sequences, resulting in the gene being inserted via attB-attP recombination (Fig. 3). Several mice models have been generated using the PhiC31 integrase system. In addition, a few studies using phiC31 recombinase were carried out in livestock, particularly cattle. However, few studies involving pigs have been reported to date [241]. Therefore, we investigated the possibility of attB-attP recombination by PhiC31 in porcine cells as well. As shown in Fig. 3, DNAs (a vector DNA; attB containing a fluorescence gene and a vector DNA; phiC31) were transfected in porcine fibroblasts with attP sequences, selected, and confirmed by genomic polymerase chain reaction. Producing pig models using PhiC31 recombination research is still in the preliminary stage. However, hopefully it can become a viable option for generating conditional transgenic pigs.
Tet-on/off. The Tet-on/off system is a powerful tool for understanding the relationships involved in gene expression. Basically, the Tet-on/off system regulates the expression of genes by tetracycline (i.e., doxycycline) indicating that the target gene expression can be on or off at specific time. Consequently, this system can be a useful model for understanding both time-dependent gene expression and specific gene expression (Fig. 4). Various studies of mice using tet-on/off models have been conducted. In addition, two pig studies for producing live transgenic piglets using this system have been carried out [1314].
In pig studies, the Tet-on system has been applied to expression of the reporter gene (fluorescence protein) [13] or the immunological gene [14]. Even though numerous tet-on/off mutant mice have been produced, progress toward applying this system to pigs has been limited. Research has demonstrated that one pig model for functional genes (RANKL and CTLA-4Ig) was dose-dependent, meaning it was regulated by doxycycline supplementation [14].
HR: HR is a classic approach to delete the endogenous gene using a selection marker. DNA for HR consists of three parts, a 5'-arm, a selection marker (such as an antibiotic resistance gene) and a 3'-arm. To clone the DNA, the long size of the left or right homology arm is needed (3 kbs greater in each arm). Consequently, it is difficult to prepare the DNA for HR and the system is very inefficient. Nevertheless, several cloned KO piglets have been generated using this method because of the importance of KO pigs. Progress in this area has been still slow because there is the limitation to increase HR efficiency in porcine primary cells. After DNA endonucleases for gene editing emerged, HR-based KO pigs were replaced with genome-editing technologies, which are briefly explained in the next section.
DNA endonuclease: Genome-editing technologies using DNA endonuclease (ZFN, TALEN and CRISPR-Cas9) have recently been developed. These technologies are efficient techniques for producing KO animals, including pigs. Prior to development of these technologies, the low efficiencies associated with HR and SCNT represented a major hurdle for generating KO pigs.
Watanabe et al. [36] were the first to report that ZFN efficiently deleted the exogenous eGFP gene from porcine somatic cells. Subsequently, the bi-allelic KO of endogenous genes (GGTA1 and CMAH) was efficiently disrupted in somatic cells. Those cells then produced KO piglets via SCNT [1217]. Even though ZFN could be efficiently applied to produce KO pigs, this technique has several disadvantages, including toxicity and off-target events [28]. Another genome-editing technology, DNA endonuclease (TALEN) and CRISPR-Cas9, has recently emerged, and it has functioned efficiently (Fig. 5). These methods have also been used to rapidly produce many KO pigs [3339]. It is expected that increasing numbers of KO and KI pigs will be produced in the near future.
In addition to the development of transgene expression and genome-editing technologies to produce mutant pig models, the improvement of SCNT has been studied consistently because it is a very practical method for generating pig models. Therefore, most transgenic pigs have been produced via SCNT with transformed cell lines via overexpression, conditional expression and KO/KI. However, SCNT-derived pig production involves epigenetic issues such as abnormalities of offspring, sudden death and low efficiency. Histone deacetylase inhibitors (HDACi) have been used to improve the SCNT approach [2743]. Additionally, in vitro maturation or culture should be improved to produce mutants pig models more efficiently [9].
As an alternative to SCNT, microinjection, which is, the direct injection of DNA into in vitro fertilized embryos, should be considered in pigs because SCNT-derived offspring exhibited epigenetic abnormalities. If this process becomes better established, then mutant pig models without abnormal epigenetic issues could be produced and grown to germ-line fertility.
Based on a literature review and our studies, we conclude that interests in the use of pig models for translation research will increase and genome engineering will become an important method to produce these models. Many mutant pigs have been developed via gene expression and genome-editing technologies. In the future, more exact gene-regulated pig models will be generated and applied to various genetic models for xenotransplantation or metabolic diseases.
Figures and Tables
 | Fig. 1Gene expression by cassette exchange via cyclic recombinase (Cre). (A) Floxed blasticidin-resistant gene by loxP and lox2272 were integrated into porcine cells. (B) Donor DNA (puromycin-linked RFP gene) and Cre recombinase were co-transfected and blasticidin gene was then exchanged. (C) Genomic polymerase chain reaction (PCR) on recombinant target genes confirmed cassette exchange by Cre recombinase. 1, DNA ladder; 2, wild type cells; 3, blasticidin integrated cells; 4, cassette exchanged cells; (−), negative control. |
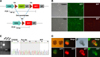 | Fig. 2Dre-rox recombination in porcine cells and embryos. (A) DNA construction and PCR-detection regions. (B) With or without Dre recombinase transfection in porcine skin fibroblasts — upper without Dre, lower with Dre. (C) Validation of DNA excision by PCR. (D) Target gene expression by Dre recombinase injection into the cloned embryos from donor cells with transfection. |
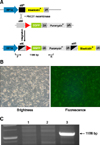 | Fig. 3Gene integration and expression by PhiC31 recombinase. (A) Porcine fibroblasts with the attP-blasticidin gene were generated. AttB-DNA and PhiC31 recombinase were co-transfected into the fibroblasts and recombination occurred. (B) After recombination, the fibroblast expressed eGFP. (C) Recombination was confirmed by genomic PCR. 1, control fibroblasts; 2, attP-transfected fibroblasts; 3, recombinated fibroblasts by PhiC31. |
 | Fig. 4Conditional gene expression with or without doxycycline. (A) Illustration of Tet-on gene expression by doxycycline. (B) RFP expression (left; with doxycycline) and non-expression (right; without doxycyline) in porcine fibroblasts after transfection of tet-on RFP vector. |
Acknowledgments
This study was financially supported by the Research Institute of Veterinary Science, IPET (No. 109023-05-5-CG000), NRF (No. 2011-0014941) and the BK21 PLUS Program for Creative Veterinary Science Research.
References
1. Anastassiadis K, Fu J, Patsch C, Hu S, Weidlich S, Duerschke K, Buchholz F, Edenhofer F, Stewart AF. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis Model Mech. 2009; 2:508–515.

2. Bi Y, Liu X, Zhang L, Shao C, Ma Z, Hua Z, Zhang L, Li L, Hua W, Xiao H, Wei Q, Zheng X. Pseudo attP sites in favor of transgene integration and expression in cultured porcine cells identified by Streptomyces phage phiC31 integrase. BMC Mol Biol. 2013; 14:20.

3. Branda CS, Dymecki SM. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004; 6:7–28.
4. Brault V, Besson V, Magnol L, Duchon A, Hérault Y. Cre/loxP-mediated chromosome engineering of the mouse
genome. Handb Exp Pharmacol. 2007; 29–48.
5. Chan AWS, Kukolj G, Skalka AM, Bremel RD. Timing of DNA integration, transgenic mosaicism, and pronuclear microinjection. Mol Reprod Dev. 1999; 52:406–413.

6. Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981; 292:154–156.

7. Flisikowska T, Kind A, Schnieke A. The new pig on the block: modelling cancer in pigs. Transgenic Res. 2013; 22:673–680.

8. Garrels W, Mátés L, Holler S, Dalda A, Taylor U, Petersen B, Niemann H, Izsvák Z, Ivics Z, Kues WA. Germline transgenic pigs by Sleeping Beauty transposition in porcine zygotes and targeted integration in the pig genome. PLoS One. 2011; 6:e23573.

9. Grupen CG. The evolution of porcine embryo in vitro production. Theriogenology. 2014; 81:24–37.
10. Gun G, Kues WA. Current progress of genetically engineered pig models for biomedical research. Biores Open Access. 2014; 3:255–264.

11. Hammer RE, Pursel VG, Rexroad CE Jr, Wall RJ, Bolt DJ, Ebert KM, Palmiter RD, Brinster RL. Production of transgenic rabbits, sheep and pigs by microinjection. Nature. 1985; 315:680–683.

12. Hauschild J, Petersen B, Santiago Y, Queisser AL, Carnwath JW, Lucas-Hahn A, Zhang L, Meng X, Gregory PD, Schwinzer R, Cost GJ, Niemann H. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc Natl Acad Sci U S A. 2011; 108:12013–12017.

13. Jin YX, Jeon Y, Lee SH, Kwon MS, Kim T, Cui XS, Hyun SH, Kim NH. Production of pigs expressing a transgene under the control of a tetracycline-inducible system. PLoS One. 2014; 9:e86146.

14. Klymiuk N, Böcker W, Schönitzer V, Bähr A, Radic T, Fröhlich T, Wünsch A, Keßler B, Kurome M, Schilling E, Herbach N, Wanke R, Nagashima H, Mutschler W, Arnold GJ, Schwinzer R, Schieker M, Wolf E. First inducible transgene expression in porcine large animal models. FASEB J. 2012; 26:1086–1099.

15. Kong Q, Hai T, Ma J, Huang T, Jiang D, Xie B, Wu M, Wang J, Song Y, Wang Y, He Y, Sun J, Hu K, Guo R, Wang L, Zhou Q, Mu Y, Liu Z. Rosa26 locus supports tissue-specific promoter driving transgene expression specifically in pig. PLoS One. 2014; 9:e107945.

16. Kragh PM, Nielsen AL, Li J, Du Y, Lin L, Schmidt M, Bøgh IB, Holm IE, Jakobsen MG, Purup S, Bolund L, Vajta G, Jørgensen AL. Hemizygous minipigs produced by random gene insertion and handmade cloning express the Alzheimer’s disease-causing dominant mutation APPsw. Transgenic Res. 2009; 18:545–558.

17. Kwon DN, Lee K, Kang MJ, Choi YJ, Park C, Whyte JJ, Brown AN, Kim JH, Samuel M, Mao J, Park KW, Murphy CN, Prather RS, Kim JH. Production of biallelic CMP-Neu5Ac hydroxylase knock-out pigs. Sci Rep. 2013; 3:1981.

18. Lai L, Kang JX, Li R, Wang J, Witt WT, Yong HY, Hao Y, Wax DM, Murphy CN, Rieke A, Samuel M, Linville ML, Korte SW, Evans RW, Starzl TE, Prather RS, Dai Y. Generation of cloned transgenic pigs rich in omega-3 fatty acids. Nat Biotechnol. 2006; 24:435–436.

19. Lai L, Park KW, Cheong HT, Kühholzer B, Samuel M, Bonk A, Im GS, Rieke A, Day BN, Murphy CN, Carter DN, Prather RS. Transgenic pig expressing the enhanced green fluorescent protein produced by nuclear transfer using colchicine-treated fibroblasts as donor cells. Mol Reprod Dev. 2002; 62:300–306.

20. Li L, Pang D, Wang T, Li Z, Chen L, Zhang M, Song N, Nie D, Chen Z, Lai L, Ouyang H. Production of a reporter transgenic pig for monitoring Cre recombinase activity. Biochem Biophys Res Commun. 2009; 382:232–235.

21. Luo W, Li Z, Huang Y, Han Y, Yao C, Duan X, Ouyang H, Li L. Generation of AQP2-Cre transgenic mini-pigs specifically expressing Cre recombinase in kidney collecting duct cells. Transgenic Res. 2014; 23:365–375.

22. Moon J, Kim S, Park H, Kang J, Park S, Koo O, da Torre BR, Saadeldin IM, Lee B. Production of porcine cloned embryos derived from cells conditionally expressing an exogenous gene using Cre-loxP. Zygote. 2012; 20:423–425.

24. Nowak-Imialek M, Kues WA, Petersen B, Lucas-Hahn A, Herrmann D, Haridoss S, Oropeza M, Lemme E, Schöler HR, Carnwath JW, Niemann H. Oct4-enhanced green fluorescent protein transgenic pigs: a new large animal model for reprogramming studies. Stem Cells Dev. 2011; 20:1563–1575.

25. Onishi A, Iwamoto M, Akita T, Mikawa S, Takeda K, Awata T, Hanada H, Perry AC. Pig cloning by microinjection of fetal fibroblast nuclei. Science. 2000; 289:1188–1190.

26. Park CG, Bottino R, Hawthorne WJ. Current status of islet xenotransplantation. Int J Surg. 2015; 23:261–266.

27. Park SJ, Park HJ, Koo OJ, Choi WJ, Moon JH, Kwon DK, Kang JT, Kim S, Choi JY, Jang G, Lee BC. Oxamflatin improves developmental competence of porcine somatic cell nuclear transfer embryos. Cell Reprogram. 2012; 14:398–406.

28. Petersen B, Niemann H. Molecular scissors and their application in genetically modified farm animals. Transgenic Res. 2015; 24:381–396.

29. Phelps CJ, Koike C, Vaught TD, Boone J, Wells KD, Chen SH, Ball S, Specht SM, Polejaeva IA, Monahan JA, Jobst PM, Sharma SB, Lamborn AE, Garst AS, Moore M, Demetris AJ, Rudert WA, Bottino R, Bertera S, Trucco M, Starzl TE, Dai Y, Ayares DL. Production of α1,3-galactosyltransferase-deficient pigs. Science. 2003; 299:411–414.

30. Sajgo S, Ghinia MG, Shi M, Liu P, Dong L, Parmhans N, Popescu O, Badea TC. Dre - Cre sequential recombination provides new tools for retinal ganglion cell labeling and manipulation in mice. PLoS One. 2014; 9:e91435.

31. Sieren JC, Meyerholz DK, Wang XJ, Davis BT, Newell JD Jr, Hammond E, Rohret JA, Rohret FA, Struzynski JT, Goeken JA, Naumann PW, Leidinger MR, Taghiyev A, Van Rheeden R, Hagen J, Darbro BW, Quelle DE, Rogers CS. Development and translational imaging of a TP53 porcine tumorigenesis model. J Clin Invest. 2014; 124:4052–4066.

32. Takahagi Y, Fujimura T, Miyagawa S, Nagashima H, Shigehisa T, Shirakura R, Murakami H. Production of α1,3-galactosyltransferase gene knockout pigs expressing both human decay-accelerating factor and N-acetylglucosaminyltransferase III. Mol Reprod Dev. 2005; 71:331–338.

33. Tan W, Carlson DF, Lancto CA, Garbe JR, Webster DA, Hackett PB, Fahrenkrug SC. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc Natl Acad Sci U S A. 2013; 110:16526–16531.

34. Uchida M, Shimatsu Y, Onoe K, Matsuyama N, Niki R, Ikeda JE, Imai H. Production of transgenic miniature pigs by pronuclear microinjection. Transgenic Res. 2001; 10:577–582.
35. Umeyama K, Watanabe M, Saito H, Kurome M, Tohi S, Matsunari H, Miki K, Nagashima H. Dominant-negative mutant hepatocyte nuclear factor 1α induces diabetes in transgenic-cloned pigs. Transgenic Res. 2009; 18:697–706.

36. Watanabe M, Umeyama K, Matsunari H, Takayanagi S, Haruyama E, Nakano K, Fujiwara T, Ikezawa Y, Nakauchi H, Nagashima H. Knockout of exogenous EGFP gene in porcine somatic cells using zinc-finger nucleases. Biochem Biophys Res Commun. 2010; 402:14–18.

37. Webster NL, Forni M, Bacci ML, Giovannoni R, Razzini R, Fantinati P, Zannoni A, Fusetti L, Dalprà L, Bianco MR, Papa M, Seren E, Sandrin MS, Mc Kenzie IF, Lavitrano M. Multi-transgenic pigs expressing three fluorescent proteins produced with high efficiency by sperm mediated gene transfer. Mol Reprod Dev. 2005; 72:68–76.

38. Wei J, Ouyang H, Wang Y, Pang D, Cong NX, Wang T, Leng B, Li D, Li X, Wu R, Ding Y, Gao F, Deng Y, Liu B, Li Z, Lai L, Feng H, Liu G, Deng X. Characterization of a hypertriglyceridemic transgenic miniature pig model expressing human apolipoprotein CIII. FEBS J. 2012; 279:91–99.

39. Whitworth KM, Lee K, Benne JA, Beaton BP, Spate LD, Murphy SL, Samuel MS, Mao J, O'Gorman C, Walters EM, Murphy CN, Driver J, Mileham A, McLaren D, Wells KD, Prather RS. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol Reprod. 2014; 91:78.
40. Yao J, Huang J, Hai T, Wang X, Qin G, Zhang H, Wu R, Cao C, Xi JJ, Yuan Z, Zhao J. Efficient bi-allelic gene knockout and site-specific knock-in mediated by TALENs in pigs. Sci Rep. 2014; 4:6926.

41. Yu Y, Tong Q, Li Z, Tian J, Wang Y, Su F, Wang Y, Liu J, Zhang Y. Improved site-specific recombinase-based method to produce selectable marker- and vector-backbone-free transgenic cells. Sci Rep. 2014; 4:4240.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download