Introduction
Cellular prion protein (PrPC) is host-encoded and ubiquitously expressed in many eukaryotic cells, especially neurons and glial cells of the mammalian central nervous system. Transmissible spongiform encephalopathies are caused by scrapie prion protein (PrPSC), which is a misfolded isoform of PrPC [18]. Although prion protein genes (Prnp) are shared by vertebrates, prion diseases are only observed in mammals [10]. A number of different functions of PrPC have been revealed in bacterial and viral diseases, including neurite outgrowth and neuroprotection, copper binding, transmembrane signaling, and antioxidant activity [4716171922]. Several investigations have focused on the role of PrPC in cell survival, apoptosis, and tumorigenesis in mammals [315212527]. We previously found that the mRNA expression level of chicken PrPC (chPrPC) was highest in the brain, and lower in the heart, lung, stomach, intestine and kidney of 15 day-old chicken embryos, similar to the results observed in other mammals. The physiology functions of chPrPC are presumably correlated with mammalian PrPC [6]. The ability of copper to bind to chicken PrP is significantly reduced when compared with humans [20]. However, other correlative functions of this ubiquitous protein in chicken are unknown.
Marek's disease (MD) is a lymphoproliferative disease of chickens caused by the MD virus (MDV) [5]. MD with widespread, metastatic T-cell lymphomas generally restricts the development of the poultry industry. However, the molecular mechanisms underlying MDV oncogenicity and pathogenicity remain unclear. Several intriguing lines of evidence indicating that PrPC may be involved in resistance to apoptosis, proliferation, and metastasis of mammalian cancer cells have recently emerged [14]. We previously used quantitative real-time PCR (qRT-PCR) to measure chPrPC mRNA expression levels in chicken embryo fibroblast cells (CEF) and MDV-infected CEF. When compared with CEF, the expression of chPrPC mRNA is markedly increased in MDV-infected CEF. These results indicate that chPrPC may play an important physiological role in MDV infection [1]. Furthermore, the expression level of chPrnp mRNA in different MD tumor tissues, adjacent nontumorous tissues, and normal chicken tissues were measured by absolute qRT-PCR analysis. Our results showed that the expression of chPrnp mRNA in MD tumor tissues is markedly increased relative to that in adjacent nontumorous tissues (p < 0.05) and normal chicken tissues (p < 0.01), and that chPrnp mRNA copies of cardiac tumor tissues, which were highest in tested tumor tissues, were present at levels 84.56 times and 559.18 times that of cardiac adjacent nontumorous tissues and normal chicken cardiac tissues. These results indicated a possible role of chPrPC in MD occurrence and development. Therefore, elucidation of the functions of chPrPC is necessary to understand the occurrence and development of MD cancer.
MD virus-transformed avian T cell line (MSB1) was obtained from MD lymphoma [2]. Our previous study demonstrated that the expression of chPrPC mRNA in MSB1 cells is 5.26 times that in splenic lymphocytes of healthy chickens. These findings indicate that the MSB1 cell line is a suitable cell model for investigating the functions of chPrPC in the occurrence and development of MD cancer in vitro. In this study, we established stable chPrPC-downregulating MSB1 cells by introducing the short interfering RNA (SiRNA) targeting chicken Prnp and investigated the effects of downregulation of chPrPC on the proliferation, invasion, migration, cell cycle, and apoptosis of MSB1 cells.
Materials and Methods
Plasmid construction
According to the pSilencer 4.1-CMV vector manual, two complementary 55 nt hairpin siRNA template oligonucleotides were designed, synthesized (Shanghai Sangon Company, China), annealed, and ligated into the pSilencer 4.1-CMV vector for the target chicken prion protein gene (chPrnp). Two reversed repeated sequences with a 19 nt target site and 2 nt overhang (5'-GAAGTTACCACAACCAGAA-3', 5'-TTCTGGTTGTGGTAACTTCCT-3') were connected by the loop (5'-TTCAAGAGA-3') in the complementary sequence, with BamH I and Hind III sites for ligation into the pSilencer 4.1-CMV vector, containing a neomycin resistance marker for the selection of stable transfectants in the presence of G418. The siRNA targeting site was derived from chPrnp cDNA (GenBank No. M95404.1). The negative control was the SiRNA sequence (5'-CGAATAGCCAGAACCAATA-3', 5'-TATTGGTTCTGGCTATTCGCT-3') with no homology to any avian gene sequence. After ligation, two plasmids were transformed into competent cells of Escherichia coli DH5α, then cultured on solid LB medium containing 50 µg/mL ampicillin at 37℃ for 24 h. Positive clones were identified by DNA sequence analysis (Shanghai Sangon Company), and the constructed recombinant and negative control plasmids were named SiRNA-3 and SiRNA-NC, respectively.
Cell culture and transfection
MSB1 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, USA) containing 10% fetal bovine serum (FBS; HyClone; GE Healthcare, Germany) and 1% penicillin–streptomycin with 5% CO2 at 37℃ [2].
MSB1 cells were transfected using Lipofectamine 2000 (Invitrogen) and recombinant plasmids (SiRNA-3 and SiRNA-NC). These transfectants were diluted 10 times and cultured in DMEM containing 800 µg/mL G418 (Invitrogen) about 48 h post-transfection for 6 days. The concentration of G418 was then decreased to 300 µg/mL for 10 days. G418-resistant clones were isolated by limited dilution in 96-well plates and transferred for enlargement. Knockdown of chPrPC clones was achieved by qRT-PCR and western blot. Finally, a stable knockdown of chPrPC MSB1 cells (MSB1-SiRNA-3) and negative control cells (MSB1-SiRNA-NC) was established.
Western blot analysis
Total proteins from MSB1, MSB1-SiRNA-3, and MSB1-SiRNA-NC cells were prepared with RIPA lysis buffer (Beyotime Institute of Biotechnology, China). After protein quantitation by UV spectrophotometer assay, equal amounts of the proteins were separated with 12% SDS–PAGE and electro-transferred onto a 0.2 micron nitrocellulose membrane. The blots were then blocked with 5% skim milk in PBST (phosphate buffered saline, pH 7.4, containing 0.05% Tween 20) for 1 h at room temperature, after which they were incubated with a rabbit polyclonal antibody against chPrPC (1 : 500, in PBST with 5% skim milk; preparation and preservation in our laboratory [13]) or a rabbit antibody against β-actin (1:1,000, to confirm equal sample loading; Sigma, USA) for 1 h at 37℃. After washing with PBST three times and incubating with HRP-conjugated goat anti-rabbit IgG (1: 5,000, in PBST with 5% skim milk; Sigma) for 1 h at 37℃, the membranes were washed three times in PBST, and the blots were stained with diaminobenzidine (DAB; PBST containing 0.5 mg/mL DAB, 0.3 g/mL mercuric chloride, and 0.33 µL/mL hydrogen peroxide). Band intensities were quantified using Image-Pro-Plus software and normalized to the quantity of β-actin.
qRT-PCR
Total RNA from MSB1, MSB1-SiRNA-3, and MSB1-SiRNA-NC cells was extracted using Trizol reagent (Invitrogen), and single stranded cDNA was synthesized using the M-MLV First Strand Kit (Invitrogen). Quantitation of mRNA levels was performed in a LightCycler480 II system (Roche, Germany) using M-MLV Platinum SYBR Green PCR SuperMix-UDG (Invitrogen) according to the manufacturer's instructions. Primers for qRT-PCR were as follows: chPrnp: forward: 5'-CCTCCGTTTGGTTTGCTG-3', reverse: 5'-GTACCGATGGTGGAGTGAGAA-3'; β-actin: forward: 5'-CTCCATACCCAAGAAAGAT-3', reverse: 5'-AATTGTGCGTGACATCAA-3'). qRT-PCR was performed as follows: 30 sec at 95℃, 30 sec at 55℃, and 30 sec at 72℃ (40 cycles). Relative chPrPC expression levels of mRNA were calculated through the double standard curve [1]. β-actin was amplified as an endogenous control.
Cell proliferation assay
Cell proliferation was determined by cell counting kit (CCK)-8 assay. Briefly, MSB1, MSB1-SiRNA-3, and MSB1-SiRNA-NC cells were plated in 96-well plates at a density of 1 × 104 cells per well in 100 µL DMEM, with three replicates for each cell line sample. After 0, 24, 48, 72, 96 or 120 h of incubation in 5% CO2 at 37℃, 10 µL of CCK-8 (Vazyme Biotech, China) solution was added to each well and cells were incubated for another 4 h. The absorbance of each well was then measured at 450 nm using a microplate reader (Bio-Rad Laboratories, USA). Blank controls were prepared by adding 100 µL DMEM to a well without cells.
Clone formation in soft agarose assay
2× DMEM with 20% FBS was mixed with 2% low melting agarose (Amresco, USA) at 37℃ and immediately placed in 24-well plates as the lower gel (0.5 mL/well). The upper gel (0.5 mL/well) was composed of equal amounts of cell suspensions (1,000 cells/mL) in DMEM (20% FBS) and 1% low-melting agarose. Three replicates for each cell line sample were set up. The plates were incubated in 5% CO2 at 37℃ until the cells in control wells had formed sufficiently large clones (the smallest clone contains more than 50 cells). The clones were then fixed in methyl alcohol and stained with 0.1% crystal violet. Finally, the plates were photographed and all clones in every well were counted under a microscope.
Apoptosis and cell cycle analys
Apoptosis was assayed using an annexin V-FITC apoptosis detection kit (Vazyme Biotech). Briefly, 1 × 106 cells were collected and washed twice with cold PBS, then resuspended in annexin-binding buffer. Annexin V–FITC (5 µL) and propidium iodide (PI) were added to the cells, after which the tubes were incubated at room temperature for 10 min in the dark. About 400 µL of annexin-binding buffer were then added to the cells, and the stained cells were analyzed using a FACSCalibur flow cytometer (BD Biosciences, USA) within 1 h.
Cells (2 × 106) were harvested and fixed in 70% ethanol for 12 h, washed twice with PBS, and then incubated with 50 mg/mL RNase A and 2.5 mg/mL PI (Sigma) for 30 min at room temperature in the dark. Finally, cells were analyzed by a FACSCalibur flow cytometer.
Cell invasion and migration assay
The invasiveness of MSB1, MSB1-SiRNA-3, and MSB1-SiRNA-NC cells was examined using Millicell Hanging Cell Culture Inserts (Millipore, USA) with an 8.0 µm pore polycarbonate membrane pre-coated with Matrigel (dilution of 1:3; Becton, Dickinson and Company, USA). Cell suspensions (4 × 104 cells) in 100 µL of serum-free DMEM were placed into the upper chambers, while the lower chambers were filled with 500 µL of DMEM supplemented with 10% FBS. Three replicates were set up for each cell line sample. After incubation in 5% CO2 at 37℃ for 18 h, cells that had invaded the lower chambers were randomly photographed and evaluated by CCK-8 assay. Migration assays were completed as described for the invasion assays using Millicell Hanging Cell Culture Inserts with an 8.0 µm pore polycarbonate membrane without Matrigel.
Statistical analysis
All experiments were carried out three times, and data are reported as the mean ± standard deviation (SD). Significant differences between samples were analyzed by one-way analysis of variance (ANOVA) at p < 0.05 or p < 0.01. All analyses were performed using the SPSS software (ver. 17.0; SPSS, USA).
Results
Efficient downregulation of chPrPC expression using SiRNA in MSB1-SiRNA-3 cells
In our previous study, three SiRNA expression plasmids (SiRNA-1, SiRNA-2, SiRNA-3) targeting chPrnp cDNA were designed, transfected into chicken fibroblast cells, and evaluated for their knockdown efficiency by qRT-PCR and Western blot at 48 h after transfection. Our results showed that 66.66% of chPrnp mRNA was knocked down by siRNA-3 compared to negative controls, while siRNA-2 and siRNA-1 knocked down 11.24% and 6.30% of the chPrnp mRNA, respectively. Similarly, siRNA-3, siRNA-2 and siRNA-1 decreased the chPrPC levels by 72.79%, 17.87% and -5.99%, respectively. These data suggested that the SiRNA-3 plasmid was most effective at decreasing chPrPC expression.
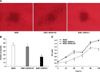
This SiRNA-3 plasmid was employed to establish stable chPrPC downregulated MSB1 cells, and G418-resistant MSB1-SiRNA-3 cells were obtained. The chPrPC mRNA and protein expression levels of MSB1, MSB1-SiRNA-3, and MSB1-SiRNA-NC cells was investigated by qRT-PCR and Western blot. In qRT-PCR experiments, the copy numbers of chPrPC and β-actin mRNA were calculated by the corresponding standard curve as follows: Ct = -3.575 log (copy Prnp) + 46.24 (R2 = 0.986); Ct = -4.218 log (copy β-actin) + 50.98 (R2 = 0.975). chPrPC relative expression was determined as previously described [1]. As shown in panel A in Fig. 1, the expression of chPrPC mRNA was significantly inhibited in MSB1-SiRNA-3 cells compared with MSB1 and MSB1-SiRNA-NC cells, with average inhibition rates of 72.83% and 85.01% (p < 0.01), respectively. Western blot revealed similar significant decreases in protein expression levels, with inhibition rates of 75.89% and 70.03% (p < 0.01; panel B and C in Fig. 1), respectively. These data indicate that the expression of chPrPC is efficiently and specifically inhibited in MSB1-SiRNA-3 cells by stable transfection with the SiRNA-3 plasmid.
Downregulation of chPrPC inhibits proliferation and clone formation of MSB1-SiRNA-3 cells
To test the effects of chPrPC expression downregulation on MSB1-SiRNA-3 cells growth, CCK-8 assays were performed to detect the proliferation of MSB1, MSB1-SiRNA-3 and MSB1-SiRNA-NC cells. Significant differences in proliferation between MSB1 and MSB1-SiRNA-NC cells were not observed. In contrast, MSB1-SiRNA-3 cell growth was significantly suppressed (panel C in Fig. 2). Clone formations in soft agarose further verified the effects of downregulation of chPrPC expression on cell growth in vitro. As expected, the number of clones of MSB1-SiRNA-3 cells (an average of 25 clones per well) generated in soft agarose was significantly reduced compared with those of MSB1 (an average of 55 clones per well) and MSB1-SiRNA-NC (an average of 51 clones per well) cells (panel B in Fig. 2). Moreover, MSB1-SiRNA-3 cells formed obvious small and localized growing clones in soft agarose (panel A in Fig. 2). These results suggest that downregulation of chPrPC not only inhibits the proliferation of MSB1-SiRNA-3 cells, but may also restrict their migration in soft agarose.
Downregulation of chPrPC induced G1 cell cycle phase arrest and apoptosis of MSB1-SiRNA-3 cells
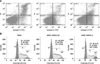
To determine if the influence of downregulation of chPrPC on cell cycle distribution or apoptosis is related to the suppression of cell growth, flow cytometric assays were performed. The population of MSB1-SiRNA-3 cells (49.31%) increased significantly (p < 0.05) at the G1 phase compared with the MSB1 (40.24%) and MSB1-SiRNA-NC (40.05%) cells (p < 0.05), while the S phase cell populations decreased. These results suggest that the cell cycle is arrested at the G1 phase by downregulation of the expression of chPrPC (panel B in Fig. 3). In addition, the total cell apoptotic rate of MSB1-SiRNA-3 cells increased markedly by 9.74% and 9.61% (p < 0.05) compared with MSB1 and MSB1-SiRNA-NC cells, respectively (panel A in Fig. 3).
Downregulation of chPrPC inhibits MSB1-SiRNA-3 cells migration and invasion
We assessed the influence of downregulation of chPrPC on the invasion and migration abilities of MSB1-SiRNA-3 cells using Millicell Hanging Cell Culture Inserts. After incubation for 18 h in the migration assay, the relative numbers of MSB1-SiRNA-3 cells that threaded the polycarbonate membrane were significantly lower than those of MSB1 and MSB1-SiRNA-NC cells based on the CCK8 assay, with average inhibition rates of 36.5% and 35.5%, respectively (p < 0.05; panels A and B in Fig. 4). Similarly, the numbers of MSB1-SiRNA-3 cells invading through the membrane of Matrigel were significantly lower than the numbers of MSB1 and MSB1-SiRNA-NC cells (panels C and D in Fig. 4), with average inhibition rates of 60.4% and 64.9%, respectively (p < 0.05). Relatively more MSB1-SiRNA-3 cells were observed in the lower chambers of the migration assay than in those of the invasion assay. These results indicate that knockdown of chPrPC inhibits MSB1-SiRNA-3 cells migration and invasion.
Discussion
RNA interference (RNAi) provides an effective technique for post-transcriptional gene knockdown through 21 to 23 nt sequence siRNA in eukaryotes [26]. To investigate gene functions and oncotherapy, researchers have constructed a considerable number of vectors that express functional siRNA when transfected into cells in vitro or in vivo [823]. Several studies have indicated the effects of PrPC on biological processes of mammalian cancer cells by employing RNAi [1215]. In the present study, we established stable chPrPC down-regulated MSB1 cells (MSB1-SiRNA-3) by introducing the SiRNA-3 plasmid targeting chicken Prnp. MSB1-SiRNA-3 cells are a useful cell model for elucidating the functions of chPrPC in the occurrence and development of MD cancer in vitro, although the expression of chPrPC in MSB1-SiRNA-3 cells was still approximately equivalent to splenic lymphocytes of healthy chickens.
While several investigations have successfully demonstrated that PrPC is involved in the resistance to apoptosis, proliferation, and metastasis of mammalian cancer cells [14], the correlative functions of chPrPC remain unknown. We examined the effects of chPrPC downregulation on cell proliferation using CCK-8 assay and clone formation in soft agarose. The results indicate that the proliferation of chPrPC-downregulated MSB1-SiRNA-3 cells was significantly suppressed compared with that of MSB1 and MSB1-SiRNA-NC cells. The following observations verified the similarity of this function with that in mammalian cell. Expression of chPrPC was positively correlated with the rate of proliferation in both the subventricular zone and dentate gyrus in adult mice [21]. Proliferation decreased in a mouse neuroblastoma cell line transfected with microRNA-targeting Prnp compared with wild-type cells [12]. Interestingly; however, the proliferation observed in the mouse brain microvascular endothelial cell line was not significantly influenced by siRNA-inducing inhibition of PrP expression compared with control cells [24]. Therefore, the expression of chPrPC in certain cells may affect proliferation through a discriminating mechanism.
To examine whether downregulation of chPrPC contributes to apoptotic promotion of MSB1-SiRNA-3 cells, flow cytometric assays were performed, and the results indicated that downregulation of chPrPC strongly increases the total cell apoptotic rate of MSB1-SiRNA-3 cells. Similar evidence has been found in mammalian cells. Downregulation of PrPC sensitizes the murine neuron derivative neuro-2a (N2a) cell line to staurosporine-induced cytotoxicity and apoptosis [27]. Overexpression of PrPC in the oral squamous cell carcinoma (HSC-2) and colon adenocarcinoma (LS 174T) cell lines confers resistance against oxidative stress-apoptosis [25]. We performed cell cycle assays, and the results showed that the cell cycle was arrested at the G1 phase by downregulating the expression of chPrPC in MSB1-SiRNA-3 cells. These findings demonstrate that downregulation of chPrPC inhibits the proliferation of MSB1-SiRNA-3 cells, at least in part, by inducing G1 cell cycle phase arrest and apoptosis. Additionally, resistance to cell apoptotosis and loss of cell cycle control are important aspects of both tumorigenesis and cancer development [911]. Taken together, these results revealed that abundant chPrPC in MD tissue is involved in tumor formation and development via anti-apoptotic and proliferation promotion.
PrPC significantly promotes the adhesive, invasive, and in vivo metastatic abilities of the gastric cancer cell lines SGC7901 and MKN45 [15]. Downregulation of PrPC suppresses migration of the mouse brain microvascular endothelial (bEND.3) cell line [24]. In our study, inhibition of MSB1-SiRNA-3 cell proliferation was also demonstrated by clone formation in soft agarose. We found that chPrPC-downregulated MSB1-SiRNA-3 cells generated smaller and fewer clones than MSB1 and MSB1-SiRNA-NC cells in soft agarose. We also observed that MSB1-SiRNA-3 cells formed evident localized clones in soft agarose, implying that downregulation of chPrPC restricts its migration. As expected, cell invasion and migration assay indicated that knockdown of chPrPC inhibits MSB1-SiRNA-3 cells migration and invasion. These findings indicate that chPrPC expression is positively correlated with the migration and invasion of MSB1 cells, similar to the results of mammalian cancer cells.
In conclusion, we established stable chPrPC downregulated MSB1 cells by introducing SiRNA-3 targeting chicken Prnp. This cell model is useful for determining the functions of chPrPC in the occurrence and development of MD cancer in vitro. Our data showed that downregulation of chPrPC inhibits MSB1 cell proliferation, migration, and invasion and induces G1 cell cycle phase arrest and apoptosis. The results indicated that chPrPC expression is positively correlated with the proliferation, migration, and invasion ability of MSB1 cells, but protects MSB1 cells from apoptosis. These findings indicate that chPrPC expression influences the formation and development of MD tumors. To the best of our knowledge, the present study is the first to provide evidence of the functions of chicken PrPC. The specific mechanism of chPrPC functions associated with cell proliferation, invasion, and apoptosis will be analyzed in future research.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download