Abstract
Previous studies have established a bovine mammary gland epithelia cells in vitro model by the adenovirus-mediated telomerase (hTERT-bMGEs). The present study was conducted to confirm whether hTERT-bMGEs were effective target cells to improve the efficiency of transgenic expression and somatic cell nuclear transfer (SCNT). To accomplish this, a mammary-specific vector encoding human lysozyme and green fluorescent protein was used to verify the transgenic efficiency of hTERT-bMGEs, and untreated bovine mammary gland epithelial cells (bMGEs) were used as a control group. The results showed that the hTERT-bMGEs group had much higher transgenic efficiency and protein expression than the bMGEs group. Furthermore, the nontransgenic and transgenic hTERT-bMGEs were used as donor cells to evaluate the efficiency of SCNT. There were no significant differences in rates of cleavage or blastocysts or hatched blastocysts of cloned embryos from nontransgenic hTERT-bMGEs at passage 18 and 28 groups (82.8% vs. 81.9%, 28.6% vs. 24.8%, 58.6% vs. 55.3%, respectively) and the transgenic group (80.8%, 26.5% and 53.4%); however, they were significantly higher than the bMGEs group (71.2%, 12.8% and 14.8%), (p < 0.05). We confirmed that hTERT-bMGEs could serve as effective target cells for improving development of somatic cell cloned cattle embryos.
The mammary gland cells of female animals are target cells for production of mammary gland bio-reactors and assessment of the function of mammary gland specific expression vectors. Unfortunately, mammary gland cells have a limited lifespan in culture of approximately 10 to 15 passages [9]. Moreover, as donor cells, extended culture for transfection and selection may induce replicative senescence and ultimately decrease somatic cell nuclear transfer (SCNT) efficiency. Some researchers are trying to use immortalized mammary cells lines [1621]; however, their effectiveness remains to be evaluated because the immortalized cells used as donor cells display developmental failure of cloned embryos [29]. In addition, most researchers have to utilize other cells in place of mammary cell lines for production of transgenic animals [1433]. In fact, these cells are unsuitable for evaluating the function of recombinant protein because expression and secretion of recombinant protein should occur in mammary cells for production of mammary gland bioreactor. Therefore, it is necessary to seek a more efficient strategy for mammary cells as nuclei donors to solve this problem. We previously reported an in vitro bovine mammary gland cells model (hTERT-bMGEs) in which an adenovirus vector carrying human telomerase reverse transcriptase (hTERT) was constructed and infected into bovine mammary gland epithelia cells (bMGEs) [10]. Our results confirmed that the adenovirus disappeared from the hTERT-bMGEs at passage 15 via successive detection of green fluorescent protein (GFP) and Western blot because of the characteristics of the unintegrated adenovirus. However, the lifespan of the hTERT-bMGEs was prolonged to more than passage 40 as a result of the initial effect of hTERT gene. The hTERT-bMGEs not only extended the cell lifespan, but also maintained the primary characteristics of the bMGEs [10]. Therefore, it provided a good cell model for subsequent research. The current study was conducted to determine whether telomerase-modified mammary epithelial cells used as nuclei donors could contribute to improvement of SCNT efficiency and enhancement of transgene expression. To accomplish this, a mammary gland specific expression vector encoding the hLYZ and GFP gene was constructed and transfected into hTERT-bMGEs to assess recombinant protein expression. Furthermore, non-transgenic and transgenic hTERT-bMGEs were used as donor cells to evaluate the efficiency of SCNT and embryonic development, respectively.
In this study, hTERT-bMGEs at passage 25 were randomly selected and cultured to detect the efficiency of gene transduction. The procedure used for culture of hTERT-bMGEs was previously described by Zheng et al. [35]. The growth medium contained 1 : 1 Dulbecco's Modified Eagle's Medium/F12 (DMEM/F12; Invitrogen, USA), plus 10% fetal calf serum (FCS; Invitrogen), 100 U/mL penicillin, 100 µg/mL streptomycin, 2 mM L-glutamine (Sigma, USA), 10 ng/mL epidermal growth factor (Invitrogen), 10 µL/mL Insulin-Transferrin-Selenium-A Supplement (100X) (Invitrogen), and 5 µg/mL insulin (Invitrogen).
A mammary gland specific expression plasmid, pEBH, was constructed as described by Yang et al. [32], in which a human cytomegalovirus (CMV) promoter drove GFP expression and a bovine β-lactoglobulin promoter drove hLYZ expression (the strategy of the vector construction was shown in Fig. 1). Additionally, the neomycin/kanamycin resistance gene was used to allow stably transfected eukaryotic cells.
The plasmid pEBH was purified using an Endo-free Plasmid Extraction Kit (Promega, USA) according to the manufacturer's instructions. The hTERT-bMGEs at passage 25 were seeded at 1 × 106 cells in 1 mL DMEM/F12 (Sigma, USA) medium supplemented with fetal bovine serum (Gibco, USA) on a 12-well tissue culture plate. The bMGEs were used as a control. Transfection was carried out by electroporation. Briefly, cells were transferred to a cold cuvette with a gap width of 0.4 cm (Bio-Rad Laboratories, USA) using two consecutive 230 V rectangular electric pulses of 20 µsec. After 72 h, the transfection efficiency of pEBH was detected using a fluorescence microscope (Eclipse TI-S; Nikon, Japan) and flow cytometry. Three independent experiments were performed.
The plasmid pEBH was transfected into hTERT-bMGEs by electroporation. The electroporation parameters were as described above. After 24 h, the transfected cells were selected using G418 (Sigma) and the positive cloning efficiency was detected. bMGEs were used as a control.
The methods were described by Yang et al. [32]. Total RNA was extracted from hLYZ-positive hTERT-bMGEs according to the instructions for Trizol Reagent (Invitrogen). A fragment of cDNA encoding hLYZ was amplified by RT-PCR using SuperScript Rerverse Transcriptase (Invitrogen). The gene fragment was synthesized from the cDNA using the following primer pairs: 5'GCACTCTGACCTAGCAGTCAACATG3' and 5'GCAGGAGAATGGCGTGAACC3'. Platinum Taq DNA Polymerase (Invitrogen) was used at 0.5 µL. The purified PCR product was cloned into the vector pMD18-T and then sequenced. Non-transgenic hTERT-bMGEs were used as a control.
The positive-hLYZ hTERT-bMGEs were obtained under G418 selection, after which the total cell lysates were subjected to SDS-PAGE in 12% polyacrylamide gel. Proteins were transferred onto PVDF membrane (Millipore, USA) and probed with anti-hLYZ (Sigma) and anti-β-actin (AC-15; Sigma) diluted 1 : 100 and 1 : 500, after which secondary antibody (Sigma) was performed at a 1 : 1,000 dilution [10]. Immunoreactive proteins were detected using an ECL detection system (Amersham, USA).
Ovaries, which were kept in compliance with the National Institutes of Health and Institutional Guidelines, were collected from a local slaughterhouse and transported to the laboratory at 35℃. The contents from follicles (2 to 8 mm in diameter) were recovered by aspirating with a 12-gauge needle. Only cumulus oocyte complexes (COCs) possessing a compact cumulus mass and evenly granulated ooplasm were pooled and washed three times with phosphate buffer saline (PBS), then washed two times with the maturation medium of COCs. COCs were cultured at 38.5℃ in an atmosphere of 5% CO2. The maturation medium comprised TCM-199 (sigma) + 10% FCS (Hyclone) + 10 µg/mL luteotropic hormone + 1 µg/mL 17 β-estradiol + 1 µg/mL follicle stimulating hormone + 1% (V : V) penicillin/streptomycin.
The 18th and 28th passage of hTERT-bMGEs used as donor cells were randomly selected to detect SCNT efficiency. The growth medium conditions were described above.
Nuclear transfer was performed according to the method reported by Ma et al. [20]. Briefly, in vitro–matured eggs with the first polar body were cultured in hepes-synthetic oviductal fluid (H-SOF) medium supplemented with 10% (v/v) FBS, 7.5 g/mL cytochalasin B (CB; Sigma) and 10 g/mL Hoechst 33342 (Sigma) at 38.5℃ for 15 min. Protrusions were then removed by aspirating micropipette with a 15 to 18 µm inner diameter, 20 to 25 µm outer diameter. A single donor cell was inserted into the perivitelline space of each egg and electrically fused using two direct pulses of 28 V for 20 ms applied two times. Fused eggs were cultured for 1 h in medium containing 0.4 mg/mL demecolcine before electroactivation, then cultured for 4 h in medium supplemented with 5 mg/mL CB. The reconstructed oocytes were activated by two direct pulses of 100 V/mm for 20 ms in 0.28 mol/L mannitol supplemented with 0.1 mM MgSO4 and 0.05 mM CaCl2. Activated eggs were cultured with 2 mmol/L 6-dimethylaminopurine (6-DMAP) in SOFaa (SOF + 2% basal medium eagle (BME) + 1% minimum essential medium (MEM) + 1 mM L-glutamine + 80 mg/mL bovine serum albumin (BSA) + 100 IU/mL penicillin + 100 µg/mL streptomycin; Sigma) medium for 4 h. The reconstructed embryos were cultured in 500 µL of SOFaa supplemented with 4 mg/mL BSA and 0.6 mM cysteine under paraffin oil on a plastic petri dish at 38.5℃. After 48 h, the egg cleavage ratio was detected. The ratio of blastocysts was then detected for 6 to 9 d.
The technique was performed according to the manufacturer's instructions (TUNEL Apoptosis Assay Kit; Roche, Switzerland). Briefly, blastocyst cells were fixed for 20 min in 4% paraformaldehyde, washed three times with PBS containing 0.1% polyvinyl alcohol (PVA), placed in 0.2% Triton X-100 for 5 min, then washed three times with PBS containing 0.1% PVA. Next, 50 µL rdTd buffer (equilibration buffer 45 µL, nucleotide mix 5 µL, rdTd enzyme 1 µL) was added and samples were held in the dark for 60 min at 37℃, followed by 2 × saline sodium citrate for 15 min at room temperature. The samples were subsequently incubated in 25 µg/mL RNase A for 30 min in the dark, after which PI was added for 30 min. Finally, the samples were covered with cover glasses and analyzed by fluorescence microscopy (Eclipse TI-S; Nikon).
To further confirm whether hTERT-bMGE could improve the efficiency for producing a mammary gland bioreactor, hLYZ-positive hTERT-bMGEs were used as donor cells for SCNT as described above. The rate of cleavage, blastocysts and hatched blastocysts were used for statistical analysis. The non-transfected hTERT-bMGEs were used as a control.
Based on the expression of GFP from pEBH, we assessed transfection efficiency by flow cytometry and fluorescence microscopy. Three independent experiments were performed (Fig. 2. shows one set of data). The transfection efficiency of the hTERT-bMGEs group was up to 36.4 ± 1% (panels A–C in Fig. 2). In contrast, the transfection efficiency of the bMGEs group was 18.6 ± 1% (panels D–F in Fig. 2). The results showed that the hTERT-bMGEs group had much higher transfection efficiency than the bMGEs groups.
Under the selection of G418, the hTERT-bMGEs group could form 78 colonies per 3 × 106 cells, while bMGEs groups could only form 15 colonies per 3 × 106 cells. The results showed that the hTERT-bMGEs group had higher colony forming efficiency than bMGEs (Table 1).
To further confirm recombinant protein expression in hTERT-bMGE, the hLYZ expression of hLYZ-positive hTERT-bMGEs was identified by RT-PCR and western blot analysis. The results showed that the hLYZ gene (850 bp) and the hLYZ protein (14.7 kDa) were both expressed in the hLYZ-positive hTERT-bMGEs, but were undetectable in non-transgenic hTERT-bMGEs (panels A and B in Fig. 3).
The indicators of a well-developed embryo are substantially synchronous cell division from the 2-, 4-, 8-, 16-cell stage, morula to blastocyst, and well-developed embryos can develop into hatched blastocysts (panels A–G in Fig. 4). Based on this, we estimated the efficiency of SCNT using the rate of cleavage, the blastocyst and hatched blastocysts. The results showed that both of the hTERT-bMGEs at passage 18 and 28 used as donor cells could develop into blastulas (panels H and I in Fig. 4), while most bMGEs at passage 10 could not (panel J in Fig. 4). In contrast, the cleavage rates of hTERT-bMGEs at passage 18 (82.8%) and 28 (81.9%) used as donor cells groups were significantly higher than those in the bMGEs group (71.2%; p < 0.05), and the blastocyst rates of the two groups (28.6% and 24.8%) were significantly higher than those of the bMGEs group (12.8%; p < 0.05; Table 2). Therefore, the results showed that the efficiency of SCNT from the hTERT-bMGEs group was much higher than that of the bMGEs group.
In general, the genetic background of the donor cells might affect the efficiency of SCNT, when foreign genes were introduced into the donor cells. Therefore, we further analyzed the development of cloned embryos from hLYZ-positive hTERT-bMGEs used as donor cells. The results indicated that hLYZ-positive hTERT-bMGEs used as donor cells could develop into a blastula (panels K and M in Fig. 4), and that most of the well-developed embryos could hatch out (panels L and N in Fig. 4). In contrast, the rates of cleavage, blastocyst development and hatched blastocysts between hLYZ-positive hTERT-bMGEs groups and non-transfected hTERT-bMGEs groups were not distinct, while those of hLYZ-positive hTERT-bMGEs groups were much higher than that of the bMGEs group (Table 2).
A high-quality embryo has the ability to successfully act as an animal mammary gland reactor. It is well known that the lower the apoptotic rate of a cloned embryo is, the better the quality becomes. Therefore, to further investigate whether the quality of hTERT-bMGEs-derived cloned embryos was high, the apoptotic rates of hTERT-bMGEs-derived cloned embryos were detected. The result showed that the apoptotic rate of the hTERT-bMGEs group (5.4 ± 1.5%) was much lower than that of the bMGEs group (15.1 ± 1.2%, p < 0.01). Thus, the lower apoptotic rate of the hTERT-bMGEs group demonstrated much higher embryo quality than that of the bMGEs group (Table 3).
The mammary gland as a bio-reactor has great commercial value. This is because the production of human recombinant protein-containing milk in transgenic farm animals offers a safe, cost-effective source [7]. Howerer, the production of transgenic mammary gland bioreactors is compromised by the low efficiency of SCNT and inefficient expression of exogenous genes. It is likely that mammary epithelial cells used as target cells have lifespans and proliferation rates in vitro that are too low to achieve the development of transgenic cloned embryos [31]. Therefore, the present study was conducted to evaluate whether telomerase could contribute to enhancement of transgene expression. The results showed that the hTERT-bMGEs group had much higher transgenic efficiency and colony efficiency than the bMGEs groups. In addition, the rate of geneticin-resistant colony formation from the hTERT-bMGEs group was higher than that in the literature [18], which might have been due to the strong proliferation ability of hTERT-bMGEs. Furthermore, the exogenous protein hLYZ maintained stable expression in positive hTERT-bMGEs by RT-PCR and western blot analysis. Therefore, positive-hLYZ hTERT-bMGEs as nuclei donors can be used to produce the transgenic cloned embryo.
We next focused on whether telomerase-modified mammary cells used as nuclei donors could contribute to the improvement of SCNT, which is a powerful tool to explore the mechanisms of reprogramming processes and biomedicine [17]. Bovine SCNT has been investigated for the production of recombinant proteins and to determine their usefulness in human disease research [192230]. Although several SCNT-derived cows have been obtained during the past few years [2122527], the overall cloning efficiency remains low, approximately 1% or less in terms of survival rate per transfer [3]. One of the reasons for this low efficiency may be insufficient epigenetic reprogramming from donor nuclei cells [2324]. Previous studies have demonstrated that incomplete reprogramming of transferred donor nuclei after SCNT caused epigenetic errors, and eventually resulted in the developmental failure of cloned embryos [131428]. Hence, it is crucial to screen and optimize suitable donor cells to improve the efficiency of SCNT [8]. Many studies have attempted to increase NT efficiency, but the results have shown small differences due to many distinct factors such as genotype [11], type of donor cell [615] and stage of donor cell [26]. The present study was designed to evaluate whether hTERT-bMGEs were effective target cells to improve the efficiency of SCNT. The results revealed better embryo quality and higher SCNT efficiency in the hTERT-bMGEs group than the control group.
Our research also investigated whether the introduction of foreign genes into the donor cell would affect the efficiency of SCNT. This is because some studies have shown a significant decline in blastocyst development rate when using transgenic cells compared to non-transgenic cells for SCNT [134]. Liu et al. [18] reported that there was no significant difference in the fusion rate and cleavage rate of cloned embryos between transgenic and non-transgenic cells, while the blastocyst formation rate of cloned embryos from a transgenic cell line was lower than those of other groups. However, our results showed that there was no difference in blastocyst formation rate, regardless of whether transgenic or non-transgenic cells were used for SCNT. These findings are consistent with those reported by Brophy [5], implying that the high proliferative activity of hTERT-bMGEs may play a role in maintaining embryo development. Moreover, generation of high-quality embryos from hTERT-bMGEs groups could be explained by extended telomere length and strong prolonged survival in hTERT-bMGEs (data not shown). This is because longer telomeres are crucial for the maintenance of genome stability, which might contribute to the improved efficiency of SCNT. Future studies should be conducted to determine the potential mechanism by which telomerase-mediated donor cells will facilitate nuclear reprogramming in hTERT-bMGEs-derived cloned embryos.
In summary, this study has provided valuable evidence showing that hTERT-bMGEs could be used in studies assessing the function of mammary gland specific expression vector and to improve the efficiency of SCNT. Moreover, the results presented herein provide additional insight into the role of telomerase activity in nuclear reprogramming of cloned embryos.
Figures and Tables
 | Fig. 1Schematic representation of a plasmid pEBH. Diagrammatic representation of the co-expression construction of pEBH contained the 5' flanking fragment (BBC5) and 3' flanking fragment (BBC3) of bovine β-lactoglobulin, human lysozyme (hLYZ), the CMV promoter, green fluorescent protein (GFP), and the neomycin/kanamycin resistance gene (Neo). |
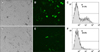 | Fig. 2Detection of transgenic efficiency of hTERT-bMGEs and bMGEs by fluorescence images and FACS analysis. hTERT-bMGEs group was a mammary gland specific expression vector (pEBH) encoding human lysozyme (hLYZ) and GFP transfer into hTERT-bMGEs, which was obtained by transfecting the adenovirus-mediated hTERT gene into bovine mammary gland epithelia cells (bMGEs), with the bMGEs used as a control. (A–C) Phase contrast, fluorescence images and FACS of bMGEs at 72 h post-transfection with plasmid pEBH. (D–F) The transfection efficiency of the hTERT-bMGEs group was much higher than that of the bMGEs group based on fluorescence detection. 100× (A, B, D and E). |
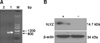 | Fig. 3(A) RT-PCR for the detection of hLYZ mRNA in hLYZ-positive hTERT-bMGEs. Lane M, 200 bp Ladder; Lane 1, the positive in hTERT-bMGEs (850 bp); Lane 2, the negative in non-transgenic hTERT-bMECs. (B) Western blot analysis of antibody specificity in hLYZ-positive hTERT-bMGEs. Expression of hLYZ in positive hTERT-bMGEs was shown by Western blot (+), but no expression of non-transgenic hTERT-bMECs was observed (-). Expression of β-actin in hLYZ-positive hTERT-bMGEs and non-transgenic hTERT-bMECs was shown by Western blot analysis. |
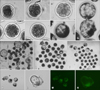 | Fig. 4The development of NT-Embryoes from hTERT-bMGEs used as donor cells. (A–G) Substantially synchronous cell divisions were shown from the 2-, 4-, 8-, 16-cell stage, stagmorula and blastocyst to the hatched blastocysts stage. (H–J) Phase contrast of blastocysts derived from hTERT-bMGEs-18, hTERT-bMGEs-28 and bMGEs-10 used as donor cells. (K and M) Phase contrast and fluorescence images of blastocyst derived from hLYZ-positive hTERT-bMGEs used as donor cells. (L and N) Phase contrast and fluorescence images of hatched blastocysts derived from hLYZ-positive hTERT-bMGEs used as donor cells. (A–N) 100×. |
Acknowledgments
This work was supported by the National Science Foundation of China (No. 31560331, 31160245 and 31460142), Natural Science Foundation of Inner Mongolia Autonomous Region of China (No. 2012MS0503), Innovation Foundation of Inner Mongolia University of Science & Technology (No. 2011NCL007) and Inner Mongolia Science & Technology Plan (No. 20140401).
References
1. Arat S, Gibbons J, Rzucidlo SJ, Respess DS, Tumlin M, Stice SL. In vitro development of bovine nuclear transfer embryos from transgenic clonal lines of adult and fetal fibroblast cells of the same genotype. Biol Reprod. 2002; 66:1768–1774.

2. Betthauser J, Forsberg E, Augenstein M, Childs L, Eilertsen K, Enos J, Forsythe T, Golueke P, Jurgella G, Koppang R, Lesmeister T, Mallon K, Mell G, Misica P, Pace M, Pfister-Genskow M, Strelchenko N, Voelker G, Watt S, Thompson S, Bishop M. Production of cloned pigs from in vitro systems. Nat Biotechnol. 2000; 18:1055–1059.

3. Bonnet-Garnier A, Veillard AC, Bed'Hom B, Hayes H, Britton-Davidian J. Assessing the quality of donor cells: karyotyping methods. Methods Mol Biol. 2015; 1222:83–99.

4. Bressan FF, Sangalli JR, Pessôa LVF, Pires PRL, Meirelles FV. Insights on bovine genetic engineering and cloning. Pesqui Vet Bras. 2013; 33:Suppl. 113–118.

5. Brophy B, Smolenski G, Wheeler T, Wells D, L'Huillier P, Laible G. Cloned transgenic cattle produce milk with higher levels of β-casein and κ-casein. Nat Biotechnol. 2003; 21:157–162.

6. Campbell KHS, Fisher P, Chen WC, Choi I, Kelly RDW, Lee JH, Xhu J. Somatic cell nuclear transfer: past, present and future perspectives. Theriogenology. 2007; 68:Suppl 1. S214–S231.

7. Carvalho AV, Reinaud P, Forde N, Healey GD, Eozenou C, Giraud-Delville C, Mansouri-Attia N, Gall L, Richard C, Lonergan P, Sheldon IM, Lea RG, Sandra O. SOCS genes expression during physiological and perturbed implantation in bovine endometrium. Reproduction. 2014; 148:545–557.
8. Firas J, Liu XD, Polo JM. Epigenetic memory in somatic cell nuclear transfer and induced pluripotency: evidence and implications. Differentiation. 2014; 88:29–32.

9. Gibson CA, Vega JR, Baumrucker CR, Oakley CS, Welsch CW. Establishment and characterization of bovine mammary epithelial cell lines. In Vitro Cell Dev Biol. 1991; 27:585–594.

10. He XY, Zheng YM, Lan J, Wu YH, Yan J, He XN, Zhang T, He YL, Zheng YL, Zhang Y. Recombinant adenovirus-mediated human telomerase reverse transcriptase gene can stimulate cell proliferation and maintain primitive characteristics in bovine mammary gland epithelial cells. Dev Growth Differ. 2011; 53:312–322.

11. Heyman Y, Chavatte-Palmer P, LeBourhis D, Camous S, Vignon X, Renard JP. Frequency and occurrence of late-gestation losses from cattle cloned embryos. Biol Reprod. 2002; 66:6–13.

12. Hodges CA, Stice SL. Generation of bovine transgenics using somatic cell nuclear transfer. Reprod Biol Endocrinol. 2003; 1:81.
13. Kang YK, Koo DB, Park JS, Choi YH, Chung AS, Lee KK, Han YK. Aberrant methylation of donor genome in cloned bovine embryos. Nat Genet. 2001; 28:173–177.

14. Kang YK, Yeo S, Kim SH, Koo DB, Park JS, Wee G, Han JS, Oh KB, Lee KK, Han YM. Precise recapitulation of methylation change in early cloned embryos. Mol Reprod Dev. 2003; 66:32–37.

15. Kato Y, Tani T, Tsunoda Y. Cloning of calves from various somatic cell types of male and female adult, newborn and fetal cows. J Reprod Fertil. 2000; 120:231–237.

16. Kung MH, Lee YJ, Hsu JT, Huang MC, Ju YT. A functional study of proximal goat β-casein promoter and intron 1 in immortalized goat mammary epithelial cells. J Dairy Sci. 2015; 98:3859–3875.

17. Kurome M, Kessler B, Wuensch A, Nagashima H, Wolf E. Nuclear transfer and transgenesis in the pig. Methods Mol Biol. 2015; 1222:37–59.

18. Liu J, Luo Y, Liu Q, Zheng L, Yang Z, Wang Y, Su J, Quan F, Zhang Y. Production of cloned embryos from caprine mammary epithelial cells expressing recombinant human β-defensin-3. Theriogenology. 2013; 79:660–666.

19. Luchsinger C, Arias ME, Vargas T, Paredes M, Sánchez R, Felmer R. Stability of reference genes for normalization of reverse transcription quantitative real-time PCR (RT-qPCR) data in bovine blastocysts produced by IVF, ICSI and SCNT. Zygote. 2014; 22:505–512.

20. Ma LB, Cai L, Li JJ, Chen XL, Ji FY. Two-staged nuclear transfer can enhance the developmental ability of goat-sheep interspecies nuclear transfer embryos in vitro. In Vitro Cell Dev Biol Anim. 2011; 47:95–103.

21. Martins LT, Gaudencio Neto S, Aguiar LH, Calderón CEM, Tavares KCS, Carneiro IS, Morais AS, Girao Neto FXA, Pinho RM, Almeida AP, Lazzarotto CR, Chies JM, Bertolini LR, Forell F, Bertolini M. 37 Effect of cell manipulation for production of transgenic cell lines on goat cloning efficiency. Reprod Fertil Dev. 2014; 27:111.

22. Matoba S, Liu Y, Lu F, Iwabuchi KA, Shen L, Inoue A, Zhang Y. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell. 2014; 159:884–895.

23. Mizutani E, Wakayama S, Wakayama T. Treatment of donor cell/embryo with different approaches to improve development after nuclear transfer. Methods Mol Biol. 2015; 1222:101–111.

24. Niemann H, Tian XC, King WA, Lee RS. Epigenetic reprogramming in embryonic and foetal development upon somatic cell nuclear transfer cloning. Reproduction. 2008; 135:151–163.

25. Onishi A, Iwamoto M, Akita T, Mikawa S, Takeda K, Awata T, Hanada H, Perry AC. Pig cloning by microinjection of fetal fibroblast nuclei. Science. 2000; 289:1188–1190.

26. Park HJ, Koo OJ, Kwon DK, Kang JT, Jang G, Lee BC. Effect of roscovitine-treated donor cells on development of porcine cloned embryos. Reprod Domest Anim. 2010; 45:1082–1088.

27. Polejaeva IA, Chen SH, Vaught TD, Page RL, Mullins J, Ball S, Dai Y, Boone J, Walker S, Ayares DL, Colman A, Campbell KHS. Cloned pigs produced by nuclear transfer from adult somatic cells. Nature. 2000; 407:86–90.

28. Santos F, Zakhartchenko V, Stojkovic M, Peters A, Jenuwein T, Wolf E, Reik W, Dean W. Epigenetic marking correlates with developmental potential in cloned bovine preimplantation embryos. Curr Biol. 2003; 13:1116–1121.

29. Shi W, Hoeflich A, Flaswinkel H, Stojkovic M, Wolf E, Zakhartchenko V. Induction of a senescent-like phenotype does not confer the ability of bovine immortal cells to support the development of nuclear transfer embryos. Biol Reprod. 2003; 69:301–309.

30. Su J, Hu G, Wang Y, Liang D, Gao M, Sun H, Zhang Y. Recombinant human growth differentiation factor-9 improves oocyte reprogramming competence and subsequent development of bovine cloned embryos. Cell Reprogram. 2014; 16:281–289.

31. Wang Y, Zhao S, Bai L, Fan J, Liu E. Expression systems and species used for transgenic animal bioreactors. Biomed Res Int. 2013; 2013:580463.

32. Yang R, Liu J, An Z, Li Z, Zhang Y. Cloning of bovine beta-casein gene and construction of mammary gland-specific expression vector. Zhongguo Sheng Wu Gong Cheng Za Zhi. 2005; 1:260–263.
33. Zakhartchenko V, Alberio R, Stojkovic M, Prelle K, Schernthaner W, Stojkovic P, Wenigerkind H, Wanke R, Düchler M, Steinborn R, Mueller M, Brem G, Wolf E. Adult cloning in cattle: potential of nuclei from a permanent cell line and from primary cultures. Mol Reprod Dev. 1999; 54:264–272.

34. Zakhartchenko V, Mueller S, Alberio R, Schernthaner W, Stojkovic M, Wenigerkind H, Wanke R, Lassnig C, Mueller M, Wolf E, Brem G. Nuclear transfer in cattle with non-transfected and transfected fetal or cloned transgenic fetal and postnatal fibroblasts. Mol Reprod Dev. 2001; 60:362–369.

35. Zheng YM, An ZX, Peng XR, Shi YQ, Zhang Y. EGFP expression in goat mammary epithelial cells. Chin J Agric Biotechnol. 2006; 3:71–74.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download