Abstract
To acquire epidemiological data on the bovine viral diarrhea virus (BVDV) and identify cattle persistently infected (PI) with this virus, 4,327 samples from Holstein dairy cows were screened over a four-year period in Beijing, China. Eighteen BVD viruses were isolated, 12 from PI cattle. Based on genetic analysis of their 5'-untranslated region (5'-UTR), the 18 isolates were assigned to subgenotype BVDV-1m, 1a, 1d, 1q, and 1b. To investigate the innate immune responses in the peripheral-blood mononuclear cells of PI cattle, the expression of Toll-like receptors (TLRs), RIG-I-like receptors, interferon-α (IFN-α), IFN-β, myxovirus (influenza virus) resistance 1 (MX1), and interferon stimulatory gene 15 (ISG15) was assessed by qPCR. When compared with healthy cattle, the expression of TLR-7, IFN-α, and IFN-β mRNA was downregulated, but the expression of MX1 and ISG-15 mRNA was upregulated in PI cattle. Immunoblotting analysis revealed that the expression of interferon regulatory factor 3 (IRF-3) and IRF-7 was lower in PI cattle than in healthy cattle. Thus, BVDV-1m and 1a are the predominant subgenotypes in the Beijing region, and the strains are highly divergent. Our findings also suggest that the TLR-7/IRF-7 signaling pathway plays a role in evasion of host restriction by BVDV.
Bovine viral diarrhea virus (BVDV), a member of the genus Pestivirus in the family Flaviviridae, is an economically important cattle pathogen with a worldwide distribution [1216]. BVDV infection can result in several cattle diseases, including respiratory and gastrointestinal infections and reproductive dysfunction [1]. BVDV causes both transient and persistent infections, which can appear as outbreaks affecting most animals or as a continuous low incidence within endemically infected herds [24].
The BVDV genome is a 12.3-12.5-kb single-stranded positive-sense RNA, which contains one large open reading frame (ORF) flanked by 5' and 3' untranslated regions (UTRs). The ORF encodes a polyprotein that is processed into structural and nonstructural proteins by viral and cellular proteases. The genetic typing of BVDV is primarily based on the 5'-UTR, Npro, and E2 regions of the genome. Epidemiological surveys ensure the effective control of BVDV [17]. For example, the genotypes and antigenic types present in a specific region must be determined so that a vaccine of the same genotype can be used [25]. In China, the occurrence of BVDV infections has been reported since 1980 [35]. However, there have been only a few reports of molecular epidemiological and phylogenetic analyses of BVDV in Chinese cattle herds.
The cytopathogenic (cp) and noncytopathogenic (ncp) biotypes of BVDV can be differentiated based on the ability of the virus to cause a cytopathic effect in tissue-cultured cells. The high numbers of cattle infected with BVDV are believed to be a consequence of the ability of ncpBVDV to establish persistent infections after in utero infection and before establishment of the host adaptive immune responses. Persistently infected (PI) cattle can act as a reservoir for the virus and are the key animals responsible for the introduction and spread of infection [13].
Although fetal infection occurs in the absence of a functional adaptive immune response, the virus must evade the host's innate immunity thereafter. Alpha/beta interferon (IFN-α/β) represents one of the first lines of defense in the innate immune system. Induction of the IFN-α/β response initially depends on the recognition of viral components by host pattern recognition receptors [21]. There is evidence that cells infected with members of the genus Pestivirus recognize viral double-stranded RNA (dsRNA) via Toll-like receptors (TLRs) and/or the retinoic acid-inducible gene I-like receptors (RLRs), including RIG-I and melanoma differentiation-associated gene 5 (MDA5) [1822]. These signals then activate the transcription factors interferon regulatory factor 3 (IRF-3) and IRF-7. Dimerized IRF-3 and/or IRF-7 translocate to the nucleus and, together with nuclear factor-κB (NF-κB), bind to both the IFNA and IFNB promoters to initiate transcription of these genes [14].
Secreted IFN-α/β triggers a signaling cascade through a common receptor, which activates the transcription of interferon-stimulated genes (ISGs), after which the products of these ISGs exert numerous antiviral effector functions [28]. The best-characterized ISG-encoded proteins with antiviral properties are 2'-5'-oligoadenylate synthetase, protein kinase R, and the myxovirus (influenza virus) resistance (MX) proteins [26].
Variable proportions of diverse immune cells are infected in PI cattle, including monocytes, T cells, and B cells. Peripheral blood mononuclear cells (PBMCs), which include B cells, T cells, monocytes, and natural killer cells, constitute a very important part of the peripheral immune system. Because PBMCs are natural reservoirs for BVDV, as well as strong producers of interferon during viral infection, and because PBMCs might play a role in dissemination of the virus throughout the body, they were selected as a suitable in vivo model in this study.
The objective of this study was to characterize the BVDV strains in the Beijing region of China and explore whether and how BVDV suppresses the IFN-α/β responses of PBMCs in PI cattle to provide new insight into the immunosuppression conferred by BVDV infection.
A total of 4,327 Holstein cattle samples (4,320 sera and seven lung tissues) were collected in the Beijing region between 2010 and 2013. None of the cattle were vaccinated against BVDV. PI cattle were tested three times with a minimum 3-week interval. Each test included BVDV antigen (antigen-capture enzyme-linked immunosorbent assay [ELISA]), genomic RNA (real-time polymerase chain reaction [RT-PCR]), and antibodies (antibody ELISA). Cattle testing positive for antigen and genomic RNA, but negative for antibody upon administration of three separate tests were considered PI [11].
Six PI cattle with no clinical symptoms and six healthy cattle were selected for this study. No other infectious pathogens (bacteria, viruses, or parasites) were isolated from the six PI cattle selected. For PI cattle, we tested viruses including BHV-1 (antibody ELISA), BRSV (antibody ELISA), PI-3 (antibody ELISA), ADV (antibody ELISA), as well as bacteria including Mycobacterium bovis (antibody ELISA), Brucella abortus (Pourquier Rose test), CBPP (antibody ELISA), MAP (antibody ELISA), and parasites including Fasciola hepatica (fecal sedimentation) and Neospora caninum (antibody ELISA). Six healthy cattle were also tested for these pathogens, and all were found to be negative. Jugular vein blood was collected from the cattle in EDTA-coated collection tubes (Becton, Dickinson and Company, UK). All animals were treated in strict accordance with the Guidelines for Laboratory Animal Use and Care of the Chinese Center for Disease Control and Prevention and the Rules for Medical Laboratory Animals (1998) of the Chinese Ministry of Health according to protocol (CAU-AEC-2013-163) approved by the Animal Ethics Committee of the China Agricultural University.
A total of 1,000 randomly selected serum samples were examined for anti-BVDV antibodies using a BVDV Total Ab Test Kit (IDEXX Laboratories, USA). Samples with S/P values higher than 0.30 were considered antibody positive. All serum samples were examined for the presence of viral Erns glycoprotein using the BVDV Antigen/SerumPlus Test Kit (IDEXX Laboratories) according to the manufacturer's instructions. Samples with a corrected optical density at a wavelength of 450 nm higher than 0.30 were classified as antigen positive.
Viral RNA was isolated from the serum samples using the EasyPure Viral DNA/RNA Kit (TransGen Biotech, China) according to the manufacturer's instructions. Viral RNA was extracted from the tissue samples with a modified Qiagen RNeasy method (Qiagen, Germany) [30]. The RNA from each sample was reverse-transcribed with a random hexamer primer using the PrimeScript RT Reagent Kit (Takara Bio, Japan) according to the manufacturer's instructions.
The PCR reaction was performed in a total volume of 50 µL containing 20 mM Tris-HCl, 50 mM KCl, 2 mM MgCl2, 200 µM dNTPs, 200 pM each of the forward and reverse primers, 4 µL of cDNA, and 1.25 U of Ex Taq DNA polymerase (Takara Bio). The reaction was heated at 94℃ for 5 min, followed by 35 cycles of 98℃ for 10 sec, 57℃ for 30 sec, and 72℃ for 30 sec, with a final elongation step at 72℃ for 1 min. After amplification, the PCR products were separated electrophoretically on 2% agarose gel in 0.5× Tris-borate-EDTA buffer. The PCR products were purified and concentrated using an Easypure Quick Gel Purification Kit (TransGen Biotech). DNA segments measuring 288 bp (fragment of the 5'-UTR) or 428 bp (fragment including the end of the 5'-UTR and part of the Npro region) were cloned into the pMD19-T Simple vector (Takara Bio) and sequenced using primers M13F and M13R (Invitrogen, USA). The 324/326 primers [32], which amplify a 288-bp fragment of the 5'-UTR, and the BD1/BD3 [33] or BD1/BD3a [4] primers, flanking a 428-bp fragment including the end of the 5'-UTR and a fragment of the Npro region were used.
The 5'-UTR and Npro sequences of the BVDVs were compared with those of isolates reported in the GenBank database. The 245-bp fragment (position in the NADL strain: nucleotides [nt] 130-374) of the 5'-UTR and the 385-bp fragment (position in the NADL strain: nt 386-770) of Npro were used for phylogenetic analysis [31]. The nucleotide sequences were proofread using the DNAMAN computer program (Lynnon Corporation, Canada) and aligned with the ClustalX analysis program (University College Dublin, Ireland). Phylogenetic trees were constructed using the MEGA version 5.2 package (Center for Evolutionary Functional Genomics, USA) based on the neighbor-joining algorithm. Evolutionary distances were calculated with the Kimura two-parameter method. The phylogenetic trees were analyzed statistically by the bootstrap method using 1,000 replicates. The corresponding GenBank accession numbers of the 5'-UTR and Npro sequences of the BVDV isolates identified in the present study are KF925503-KF925525.
PBMCs were prepared with Ficoll gradient centrifugation. Briefly, 10 mL of Ficoll-Hypaque (TBDscience, China) was stratified under 10 mL of diluted peripheral blood (anticoagulated blood mixed with an equal volume of phosphate-buffered saline [PBS]) and centrifuged at 400 ×g for 20 min at 22℃ according to the manufacturer's instructions. The recovered PBMCs were washed three times with cell culture medium, after which the samples were stained with Trypan blue (Sigma-Aldrich, USA) at a concentration of 0.4 mg/mL and the living cells were counted under a microscope (400×, TS-100; Nikon, Japan). Freshly isolated bovine PBMCs were used for all subsequent experiments.
Total RNA was extracted from PBMCs with TRIzol Reagent (Invitrogen). The final RNA was eluted in an appropriate amount of diethyl pyrocarbonate-treated water. The integrity of the RNA extracted from each sample was confirmed by agarose gel electrophoresis, with ethidium bromide staining and visualization under UV light. The amount of RNA extracted was determined and its purity was verified (OD260/OD280 absorption ratio > 1.9) with a NanoDrop-2000c Spectrophotometer (Thermo Scientific, USA). Complementary DNA was synthesized from the total RNA using M-MLV Reverse Transcriptase (Promega, USA), then stored at -80℃ until real-time PCR analysis. Quantitative real-time PCR was performed with the SYBR Premix DimerEraser (Takara Bio) and an ABI 7500 Real-time PCR System (Applied Biosystems, USA). The sequences of the primers are listed in Table 1. Each system was reevaluated and optimized for the protocol used, and all reagents were titrated for optimal performance using total RNA isolated from bovine PBMCs. To quantify the relative mRNA expression, the Ct values of the target genes were normalized to the Ct values of the housekeeping gene encoding β-actin using the 2-ΔΔCt method and the results were presented as fold changes. The PCR cycling parameters were as follows: initial denaturation at 95℃ for 30 sec, then 40 cycles at 95℃ for 5 sec, 55℃ for 30 sec, and 72℃ for 15 sec. Each PCR run was followed by a melting-curve stage and all melting curves displayed a single peak, consistent with the presence of a single amplified product. All reactions were performed in quadruplicate.
Isolated PBMCs were washed twice with ice-cold PBS, and whole-cell lysates were prepared with RIPA buffer (Sigma-Aldrich) according to the manufacturer's instructions. The protein concentrations in the supernatants were determined with the BCA Protein Assay kit (CoWin Biotech, China). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) 5× loading buffer (CoWin Biotech) was added to the lysates. The lysates were then heated to 100℃ for 5 min, after which 20 µg of protein was loaded into each well of a 10% SDS-PAGE gel. The resolved proteins were transferred electrophoretically onto polyvinylidene difluoride membrane (Millipore, USA) and blocked with 5% nonfat milk (Becton, Dickinson and Company). The membrane was subsequently incubated with a rabbit anti-human IRF-3 polyclonal antibody (diluted 1:500; BBI, China) or rabbit anti-human IRF-7 polyclonal antibody (diluted 1:500; Proteintech, USA). Following overnight incubation at 4℃, the blots were washed, exposed to secondary antibody (Peroxidase-Affinipure Donkey Anti-rabbit IgG [H+L], diluted 1:5,000; Proteintech) for 50 min at 37℃, and finally detected using an ECL WB kit (CoWin Biotech). The β-actin fragment present in each sample was used as the internal control. Each band was analyzed using the Quantity One version 4.6.2 densitometric software (Bio-Rad Laboratories, USA). The results are presented as the individual ratios of the IRF-3 and IRF-7 band intensities to the intensity of the β-actin band.
The prevalence of BVDV antibody-positive animals was 93.4% when 1,000 randomly selected serum samples were tested. Eighteen of 4,327 samples were confirmed to be BVDV-antigen-positive using an antigen-capture ELISA combined with RT-PCR. Of the BVDV-positive cattle, 13 had no or very minor symptoms, three experienced abortion, one had pneumonia, and one had diarrhea. Notably, 12 cattle were confirmed as PI. The positive samples were then processed for viral isolation to generate viral stocks. Cell culture indicated that all isolates belonged to the ncp biotype. Descriptions of the 18 isolates are given in Table 2.
Based on genetic analysis of the 5'-UTR sequences, the 18 isolates were assigned to the subgenotypes BVDV-1m (six isolates), 1a (five isolates), 1d (three isolates), 1q (three isolates), and 1b (one isolate). The BVDV-1m and 1a isolates showed 95.2% to 99.7% and 99.0% to 99.8% internal nucleotide homology, respectively, in the 5'-UTR region of the virus. Although the three isolates of BVDV-1d were from three different districts/counties, they shared high levels of nucleotide homology (99.7%-100%). The BVDV-1q strains isolated in the present study had nucleotide sequences highly homologous (98.6%-100%) to those of isolates found in swine. Thirty-seven BVDV reference strains were retrieved from GenBank for 5'-UTR analysis (panel A in Fig. 1). A phylogenetic tree was also constructed based on the Npro regions of the five strains. Twenty-two BVDV reference strains were retrieved from GenBank and added to the Npro dataset (panel B in Fig. 1).
The expressions of TLR-2, TLR-3, TLR-4, TLR-7, TLR-8, RIG-I, and MDA5 mRNA in the PBMCs of healthy and PI cattle were determined. TLR-7 mRNA expression was significantly downregulated (p < 0.001) in PBMCs from PI cattle compared with that in PBMCs from healthy cattle, whereas the expression of other TLRs, including TLR-2, TLR-3, TLR-4, and TLR-8, did not differ significantly between healthy and infected cattle. The expression of neither RIG-I nor MDA5 mRNA differed markedly between groups (Fig. 2).
We compared the mRNA expression of IFN-α, IFN-β, and MX1 in PBMCs from PI cattle and healthy cattle. IFN-α (p < 0.001) and IFN-β (p < 0.001) mRNA were lower in the PBMCs from PI cattle than in those from healthy cattle. However, MX1 mRNA expression was increased (p = 0.002) in the PBMCs from PI cattle. We also monitored the expression of ISG-15 mRNA, which was higher in the PBMCs from PI cattle than in those from healthy cattle (p < 0.001; Fig. 3).
To determine if downregulation of IFN-α/β expression could be attributed to the downregulated expression of IRFs, we analyzed the levels of IRF-3 and IRF-7 by immunoblotting. Both IRF-3 (p = 0.003) and IRF-7 (p = 0.032) expression was downregulated in PBMCs from PI cattle compared with their expression in healthy cattle (Fig. 4).
In this study, 12 of 18 BVDV strains were isolated from PI cattle. PI animals ensure the persistence of the virus in the host population because they can shed large amounts of virus into the environment throughout their lives. Therefore, herds that contain PI animals usually have continuously high levels of anti-BVDV antibodies. In this study, the prevalence of antibody-positive animals reached 93.4%. This situation might have resulted from PI cattle. However, for young cattle it might be caused by colostrum without prior infection.
Genotypic analyses are useful in epidemiological studies of
viruses because the origins of new outbreaks can be traced. The phylogenetic data obtained in the present study indicate that BVDV-1m and 1a are the predominant subgenotypes in cattle herds in the Beijing region. The first BVDV-1m strain, ZM-95, was initially isolated from swine herds in Inner Mongolia in 1995 [34]. Accumulating evidence has since strongly supported the idea that BVDV-1m is emerging as the predominant subgenotype in China [68]. The high nucleotide homology among BVDV-1m strains, including those from current and previous reports, implies that they share the same origin [635]. BVDV-1a is the predominant subgenotype in many countries throughout the world. Previous studies have shown that this subgenotype prevails in swine herds in Shanghai and in Bactrian camel herds in Xinjiang [68]. There are two possible explanations for the association between the BVDV-1a genotype and cattle infections in the Beijing region identified in this study. On one hand, some dairy cattle were imported from Australia, where the BVDV-1a subgenotype is prevalent [25]. On the other hand, the frequent domestic circulation of livestock animals offers an opportunity for cross-species infections with BVDV.
Three BVDV-1d viruses were isolated in the present study. A recent study reported that BVDV-1d exists in cattle herds in Northwestern China [10], and our results indicate the increasing prevalence of genotype 1d in China. In the present study, BJ1022, BJ1123, and BJ1301 clustered within BVDV-1q in the 5'-UTR-based analysis. This subgenotype was first identified in swine herds in Jilin, Shandong, Fujian, and Zhejiang Provinces of China in 2012 [6]. In a more recent study, it was also found in camels in Xinjiang, and initially designated '1q' [8]. Only BJ1021 was grouped in subgenotype BVDV-1b in the present study. The 99.6% nucleotide homology between BJ1021 and TJ0802 (isolated from Tianjin) suggests that they share the same origin [35].
PI animals are the key factor in the spread of this virus in dairy cattle herds. As a prerequisite for persistence, BVDV infections must first occur during early development of the fetus, and the virus must evade the innate immune response. It seems that more than one mechanism is used to inhibit the induction of IFN-α/β during ncpBVDV infection in vitro. First, structural protein Erns inhibits type I IFN by degrading viral dsRNA [19]. Second, nonstructural protein Npro blocks IFN-α/β expression by targeting IRF-3 for degradation by polyubiquitination [13]. However, the mechanism of innate immune evasion by ncpBVDV has not been fully clarified. For example, current studies cannot explain how ncpBVDV inhibits IFN-α expression. The inhibition of IFN-α expression might not simply result from reduced IRF-3, because the IRF-7 homodimer also promotes the transcription of IFN-α [23]. In addition, although both the RNase activity of Erns and the degradative activity of Npro are believed to be involved in evasion of the type I IFN response by BVDV in vitro, the details regarding the mechanism in vivo are largely unknown. Here, we have provided evidence that BVDV inhibits IFN-α/β response in PBMCs in PI cattle by inhibiting the expression of TLR-7, IRF-3, and IRF-7.
The TLRs, including TLR-3, TLR-7, and TLR-8, and RLRs, such as RIG-I and MDA5, act as initial sensors of RNA viral infection and trigger signal transduction cascades that induce the expression of type I IFN. TLR-3, RIG-I, and MDA5 respond to dsRNA, which is produced during the replication of many RNA viruses, whereas TLR-7 and TLR-8 are implicated in the recognition of virus-derived single-stranded RNA [23]. In our study, PBMCs from PI cattle showed significantly lower TLR-7 mRNA expression than those of healthy cattle, whereas the other receptors tested here did not differ significantly between healthy and infected cattle. TLR-7 is primarily expressed by plasmacytoid dendritic cells and is critical for the efficient control of acute and persistent viral infections. Downregulated TLR-7 expression has been reported during hepatitis C virus infection and is a potential mechanism for its evasion of the host's innate immune system [5]. A possible mechanism underlying our observation is that ncpBVDV interferes with the TLR-7 gene by reducing its stability during its transcriptional regulation in the PBMCs of PI cattle [5].
The transcription factors IRF-3 and IRF-7 are considered master regulators of type I IFN induction. It has been shown that during ncpBVDV infections in vitro, Npro can induce the long-term downregulation of IRF-3 expression by inducing its polyubiquitination and proteasomal degradation [13]. In the present study, both IRF-3 and IRF-7 expression were reduced in the PBMCs of PI cattle relative to their expression in healthy cattle. This observation indicates that ncpBVDV evades the innate immune response in vivo, not only via IRF-3, but also via IRF-7. Certain cells, particularly antigen-presenting cells, constitutively express IRF-7, which potentiates them for rapid IFN-α production [27]. Therefore, a reduction in IRF-7 might result when monocytes (a type of PBMCs) are disabled because the tropism of BVDV for monocytes can compromise their capacity to function normally [9]. Indeed, it was reported that Npro of classic swine fever virus (CSFV), which also belongs to the genus Pestivirus, might induce the degradation of IRF-7 in vitro [7]. In this report, the author hypothesized that Npro of CSFV might affect the localization of IRF-7, and that reduced translocation of IRF-7 might modulate the proteasomemediated IRF-7 turnover [7]. However, the underlying mechanism for the effects of BVDV on IRF-7 expression in antigen-presenting cells in cattle remains to be determined.
In the present study, the PBMCs from all PI cattle showed reduced IFN-α/β mRNA expression. Based on these findings, reduced expression of IFN-α/β mRNA results from the downregulated expression of IRF-3 and IRF-7 because these proteins play essential roles in ensuring the efficient and diverse transcription of the type I IFN genes in the antiviral response [15]. Interestingly, although IFN-α/β and IRF-3/7 were significantly reduced, PBMCs from PI cattle showed increases in MX1 and ISG-15 mRNA expression. The increased expression of ISG mRNA might result from the upregulated expression of IFN-γ or other IRFs (such as IRF-9), which also induce ISG expression [20]. Of course, as PBMCs are a highly heterogeneous cell population, the increase in mRNA of MX1 and ISG-15 might result from one cell type. Therefore, it seems that ncpBVDV infection does not totally suppress the innate immune responses that control nonself pathogens in PI cattle [29].
In conclusion, the results of this study indicate that BVDV infections are ubiquitous in cattle herds in the Beijing region of China and that PI cattle contribute to this situation. Moreover, ncpBVDV can evade the innate immune response of the host by downregulating TLR-7, IRF-3, and IRF-7 expression in PI cattle, consequently reducing the production of IFN-α/β.
Figures and Tables
 | Fig. 1Phylogenetic analysis of 245 nucleotides of the 5'-UTR or 385 nucleotides from the Npro genomic region in diverse strains of bovine viral diarrhea virus (BVDV). The 5'-UTR fragment (A) was determined in 18 of the analyzed strains, and a fragment of the Npro region (B) was determined in another five strains. Numbers over branches indicate the percentage of one thousand bootstrap replicates. The strains sequenced in this study are labeled with black triangles. GenBank accession nos. of reference strains in the 5'-UTR region were as follows: ZM-95 (AF526381), Shihezi148 (EU159700), Manasi (EU159702), SH1051 (JN248740), NX0803 (GU120254), DG07 (GU120250), TJ06 (GU120246), BJ0701 (GU120247), JS10116 (JN248734), camel-6 (KC695810), SD0803 (JN400273), FJ1003 (JN248728), ZJ1003 (JN248744), SH0710 (JN248738), SH1060 (JN248741), NADL (AJ133738), CP7 (U63479), KA07 (GQ495691), TJ0802 (GU120256), TrangieY546 (AF049222), KA01 (GQ495677), KB01 (GQ495676), 10-84 (AF298054), IT99-7101 (AJ318618), cell-con-1 (KC695816), F-Au (AF298065), J-Au (AF298067), W-Au (AF298073), A-Au (AF298064), L-Au (AF298069), G-Au (AF298066), KS86-1ncp (AB078950), Rebe (AF299317), SuwaCp (AF117699), 06z127 (DQ973181), XJ-04 (FJ527854), and 890 (U18059). Accession nos. of the BVDV reference strains in the Npro region are as follows: NADL (AJ133738), SD-1 (M96751), Deer-NZ1 (U80903), Shitara/01/05 (AB359926), F-Au (AF287284), 3186V6 (AF287282), J-Au (AF287286), W-Au (AF287290), A-Au (AF287283), L-Au (AF287287), G-Au (AF287285), 24/15 (AF287280), DeerGB1 (U80902), TR27 (EU163975), ZM-95 (AF526381), TJ0801 (GU120262), IS25CP/01 (AB359931), Shitara/02/06 (AB359930), SoCP/75 (AB359929), 871 (AF144462), Osloss (M96687), and BVDV-2 strain 890 (U18059). |
 | Fig. 2Expression of mRNA encoding (A) TLR-2, (B) TLR-3, (C) TLR-4, (D) TLR-7, (E) TLR-8, (F) RIG-I, and (G) MDA5 in peripheral blood mononuclear cells (PBMCs) collected from healthy cattle and PI cattle based on quantitative real-time PCR. Data presented are the means ± SEM (n = 6 per group). P < 0.05 is considered statistically significant. |
 | Fig. 3Expression of mRNA encoding (A) IFN-α, (B) IFN-β, (C) MX1, and (D) ISG-15 in PBMCs collected from healthy and PI cattle based on quantitative real-time PCR. Data presented are the means ± SEM (n = 6 per group). P < 0.05 is considered statistically significant. |
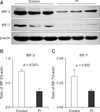 | Fig. 4Immunoblotting detection of IRF-3 and IRF-7 in PBMCs from healthy and PI cattle. (A) Representative panels of IRF-3 and IRF-7 are shown. The expression of (B) IRF-3 and (C) IRF-7. Protein lysates were prepared with protease inhibitors and the expression of IRF-3 and IRF-7 was analyzed by immunoblotting, detected with enhanced chemiluminescence, and then analyzed with the Quantity One densitometric software. Simultaneous staining of β-actin on the same membrane was used as a control for protein loading in the individual lanes. Results are presented as the ratio of the intensity of the IRF-3 or IRF-7 band to the intensity of the β-actin band. Data presented are the means ± SEM (n = 6 per group). P < 0.05 is considered statistically significant. |
References
1. Baker JC. The clinical manifestations of bovine viral diarrhea infection. Vet Clin North Am Food Anim Pract. 1995; 11:425–445.

2. Berke IC, Yu X, Modis Y, Egelman EH. MDA5 assembles into a polar helical filament on dsRNA. Proc Natl Acad Sci U S A. 2012; 109:18437–18441.

4. Booth RE, Thomas CJ, El-Attar LM, Gunn G, Brownlie J. A phylogenetic analysis of Bovine viral diarrhoea virus (BVDV) isolates from six different regions of the UK and links to animal movement data. Vet Res. 2013; 44:43.

5. Chang S, Kodys K, Szabo G. Impaired expression and function of toll-like receptor 7 in hepatitis C virus infection in human hepatoma cells. Hepatology. 2010; 51:35–42.

6. Deng Y, Sun CQ, Cao SJ, Lin T, Yuan SS, Zhang HB, Zhai SL, Huang L, Shan TL, Zheng H, Wen XT, Tong GZ. High prevalence of bovine viral diarrhea virus 1 in Chinese swine herds. Vet Microbiol. 2012; 159:490–493.

7. Fiebach AR, Guzylack-Piriou L, Python S, Summerfield A, Ruggli N. Classical swine fever virus Npro limits type I interferon induction in plasmacytoid dendritic cells by interacting with interferon regulatory factor 7. J Virol. 2011; 85:8002–8011.

8. Gao S, Luo J, Du J, Lang Y, Cong G, Shao J, Lin T, Zhao F, Belák S, Liu L, Chang H, Yin H. Serological and molecular evidence for natural infection of Bactrian camels with multiple subgenotypes of bovine viral diarrhea virus in Western China. Vet Microbiol. 2013; 163:172–176.

9. Glew EJ, Carr BV, Brackenbury LS, Hope JC, Charleston B, Howard CJ. Differential effects of bovine viral diarrhoea virus on monocytes and dendritic cells. J Gen Virol. 2003; 84:1771–1780.
10. Gong X, Cao X, Zheng F, Chen Q, Zhou J, Yin H, Liu L, Cai X. Identification and characterization of a novel subgenotype of bovine viral diarrhea virus isolated from dairy cattle in Northwestern China. Virus Genes. 2013; 46:375–376.

11. Hanon JB, Van der Stede Y, Antonissen A, Mullender C, Tignon M, van den Berg T, Caij B. Distinction between persistent and transient infection in a bovine viral diarrhoea (BVD) control programme: appropriate interpretation of real-time RT-PCR and antigen-ELISA test results. Transbound Emerg Dis. 2014; 61:156–162.

12. Heuer C, Healy A, Zerbini C. Economic effects of exposure to bovine viral diarrhea virus on dairy herds in New Zealand. J Dairy Sci. 2007; 90:5428–5438.

13. Hilton L, Moganeradj K, Zhang G, Chen YH, Randall RE, McCauley JW, Goodbourn S. The NPro product of bovine viral diarrhea virus inhibits DNA binding by interferon regulatory factor 3 and targets it for proteasomal degradation. J Virol. 2006; 80:11723–11732.

14. Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem. 2007; 282:15325–15329.

15. Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005; 434:772–777.

16. Houe H. Epidemiological features and economical importance of bovine virus diarrhoea virus (BVDV) infections. Vet Microbiol. 1999; 64:89–107.

17. Houe H. Epidemiology of bovine viral diarrhea virus. Vet Clin North Am Food Anim Pract. 1995; 11:521–547.

18. Hüsser L, Alves MP, Ruggli N, Summerfield A. Identification of the role of RIG-I, MDA-5 and TLR3 in sensing RNA viruses in porcine epithelial cells using lentivirus-driven RNA interference. Virus Res. 2011; 159:9–16.

19. Iqbal M, Poole E, Goodbourn S, McCauley JW. Role for bovine viral diarrhea virus Erns glycoprotein in the control of activation of beta interferon by double-stranded RNA. J Virol. 2004; 78:136–145.

22. Lee SW, Park Y, So T, Kwon BS, Cheroutre H, Mittler RS, Croft M. Identification of regulatory functions for 4-1BB and 4-1BBL in myelopoiesis and the development of dendritic cells. Nat Immunol. 2008; 9:917–926.

23. Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005; 5:375–386.

24. Potgieter LN. Immunology of bovine viral diarrhea virus. Vet Clin North Am Food Anim Pract. 1995; 11:501–520.

25. Ridpath JF, Fulton RW, Kirkland PD, Neill JD. Prevalence and antigenic differences observed between Bovine viral diarrhea virus subgenotypes isolated from cattle in Australia and feedlots in the southwestern United States. J Vet Diagn Invest. 2010; 22:184–191.

26. Sadler AJ, Williams BRG. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008; 8:559–568.

27. Schmid S, Mordstein M, Kochs G, García-Sastre A, tenOever BR. Transcription factor redundancy ensures induction of the antiviral state. J Biol Chem. 2010; 285:42013–42022.

28. Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011; 472:481–485.

29. Schweizer M, Mätzener P, Pfaffen G, Stalder H, Peterhans E. "Self" and "nonself" manipulation of interferon defense during persistent infection: bovine viral diarrhea virus resists alpha/beta interferon without blocking antiviral activity against unrelated viruses replicating in its host cells. J Virol. 2006; 80:6926–6935.

30. Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, Atkins D, Wang Y. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005; 11:7234–7242.

31. Vilcek S, Durkovic B, Kolesárová M, Greiser-Wilke I, Paton D. Genetic diversity of international bovine viral diarrhoea virus (BVDV) isolates: identification of a new BVDV-1 genetic group. Vet Res. 2004; 35:609–615.

32. Vilcek S, Herring AJ, Herring JA, Nettleton PF, Lowings JP, Paton DJ. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch Virol. 1994; 136:309–323.

33. Vilcek S, Paton DJ, Durkovic B, Strojny L, Ibata G, Moussa A, Loitsch A, Rossmanith W, Vega S, Scicluna MT, Paifi V. Bovine viral diarrhoea virus genotype 1 can be separated into at least eleven genetic groups. Arch Virol. 2001; 146:99–115.

34. Wang X, Tu C, Li H, Jin K, Xuan H, Chang G, Sun H, Zhu W, Fei E, Yin Z. Pigs naturally infected by bovine diarrhea virus present signs resembling hog cholera. Chin J Vet Sci. 1996; 16:341–345.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download