Abstract
Amplification of the 16S rRNA gene from a blood sample obtained from a dog in southeastern Brazil was used to confirm a naturally acquired Ehrlichia (E.) canis infection. Following isolation and culturing of the new bacterial strain called Uberlândia, partial sequences of the dsb and p28 genes were obtained. The dsb partial sequence of the novel strain was 100% similar to dsb gene sequences of E. canis obtained from different geographic areas around the world. Conversely, the p28 partial sequence for the E. canis Uberlândia strain differed at several nucleotides from other sequences available in GenBank. To confirm the antigenic profile of the Uberlândia strain, an indirect immunofluorescence assay against E. canis antigens was performed using dog sera collected from two different areas in Brazil (Uberlândia and São Paulo). The results suggest that both antigens were able to identify animals seropositive for E. canis in Brazil since these Brazilian strains appear to be highly conserved.
Ehrlichia (E.) canis is the primary etiologic agent of canine monocytic ehrlichiosis (CME) in domestic dogs worldwide wherever the main vector, the brown dog tick Rhipicephalus sanguineus, is prevalent. In Brazil, ehrlichiosis was first reported in the southeast part of the area [10]. Currently, the disease is found throughout the country and considered endemic in many regions [29,34].
Cell culture isolation (CCI) allows for further characterization of cytotoxic agents for a sensitive serologic test and can help identify geographic-specific antigens [33]. Until now, only three isolates of E. canis from different parts of Brazil, Rio de Janeiro city [32], Jaboticabal [2], and São Paulo city [1], have been propagated in vitro. Most molecular epidemiology studies of E. canis have focused on the 16S ribosomal RNA gene (16S rRNA). Consequently, much less is known about the other genes of this microorganism. Molecular characterization of 16S rRNA has provided information about strain diversity and suggests a high level of conservation [16,24]. Little is known about the disulfide bond formation protein gene (dsb) or the role of the inter- and intramolecular disulfide bonds in the overall Ehrlichia cell envelope structure. However, these genes are functionally conserved among bacteria, and recombinant Ehrlichia dsb proteins are found in the periplasm of E. chaffeensis and E. canis [20]. Thus, the dsb gene is a useful target for diagnosing infection by medically important ehrlichial species [12].
The p28 protein acts as an adhesin in the outer Ehrlichia membrane [25]. It has been detected in vitro [21] and in the vertebrate host [27]. The gene encoding this protein may be useful as a diagnostic antigen and the eventual creation of a vaccine against ehrlichioses [11,30].
Although many studies have been conducted, little is known about the genetic diversity of E. canis strains worldwide. The present study was therefore performed to isolate and partially characterize the genetic features of a novel E. canis Brazilian strain. Additionally, the efficacy of different antigens from Brazil for diagnosing CME was evaluated.
The experimental protocol (n°. 017/09) was approved by the Experimental Animal Ethics Committee of the Federal University of Uberlândia (Brazil).
A 5-year-old male poodle was admitted to the Veterinary Hospital of the Federal University of Uberlândia in June 2009. The animal had a history of tick exposure, and presented apathy, fever (39℃), and pale mucous membranes during physical examination. Complete blood cell counts revealed that the dog suffered from thrombocytopenia [platelet count: 33 × 103/µL (reference values: 200~500 × 103/µL)], anemia [hematocrit: 11.8% (reference values: 37~55%); hemoglobin: 3.7 g/dL (reference values: 12~18 g/dL); erythrocytes count: 1.8 × 106/µL (reference values: 5.5~8.5 × 106/µL)], and leukocytosis with a left shift [leucocytes count: 20.6 × 103/µL (reference values: 5~10 × 103/µL)]. Blood sample (3 mL) was collected aseptically into vacutainer tubes containing EDTA and heparin for PCR and CCI, respectively.
The mononuclear cell fraction was isolated from heparinized blood with Histopaque 1083 (Sigma-Aldrich, USA) and used to inoculate a monolayer of DH82 cells (ATCC no. CRL-10389) according to Aguiar et al. (2008). Briefly, leukocytes were isolated by overlaying the buffy coat on Histopaque 1083 (Sigma-Aldrich, USA), and the interface containing the leukocyte fraction was collected and resuspended in 5 ml of Dulbecco's Modified Eagle's medium (Sigma-Aldrich, USA) supplemented with 10% heat-inactivated bovine calf serum (Hyclone Laboratories, USA). The leukocyte suspension was transferred to a 25 cm2 flask culture and incubated at 37℃ in a 5% CO2 atmosphere. After 24 hrs, 2 ml of fresh culture medium was added and the cells harvested 24 hrs later and added to a monolayer of uninfected DH82 cells (provided by Marcelo B. Labruna, University of São Paulo) in a 25 cm2 flask. The culture was maintained under the same conditions as above, except that the bovine calf serum was reduced to 2.5%, and the culture medium was partially (20%) replaced every 2~3 days. Cell cultures were monitored once a week for the presence of ehrlichial morulae by Diff-Quik staining according to the manufacturer's instructions (Laborclin Produtos para Laboratórios, Brazil) and ehrlichial DNA by nested PCR.
DNA was extracted from either 200 µl of canine peripheral blood or from pelleted DH82 cell cultures infected and non-infected using a DNAeasy Tissue Kit (Qiagen, Germany). DNA was eluted with ultrapure water from Milli-Q Integral System (Merck Millipore, USA), quantified with a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, USA), and stored at -20℃.
The 16S rRNA gene was amplified from peripheral dog blood and DH82 cells. Two pairs of primers were used for the nested PCR, ECC (5'-AGAACGAACGCTGGCGGCAAGC-3') combined with ECB (5'-CGTATTACCGCGGCTGCTGGCA-3'), to amplify a product approximately 478 bp in size. The product was tested with inners primers, ECAN5 (5'-CAATTATTTATAGCCTCTGGCTATAGGA-3') and HE3 (5'-TATAGGTACCGTCATTATCTTCCCTAT-3'), which amplify a 365-bp fragment [23]. To analyze the dsb gene, a 409-bp fragment was amplified with the dsb-330 forward (5'-GATGATGTCTGAAGATATGAAACAAAT-3') and dsb-728 reverse (5'-CTGCTCGTCTATTTTACTTCTTAAAGT-3') primers [12]. To examine the p28 gene, the forward 793 (5'-GCAGGAGCTGTTGGTTACTC-3') and reverse 1,330 (5'-CCTTCCTCCAAGTTCTATGCC-3') primers were used to amplify a 518-bp fragment [21]. Each reaction contained 25 pmol of each primer, 1.25 U of Platinum Taq DNA Polymerase (Invitrogen, USA), PCR buffer (50 mM KCl and 20 mM Tris-HCl) (Invitrogen, USA), 2 mM MgCl2 (Invitrogen, USA), dNTP mixture (0.25 mM each) (Invitrogen, USA), 100~200 ng of DNA template, and ultrapure water from Milli-Q Integral System (Merck Millipore, USA) in a final volume of 50 µL. PCR was performed in an automatic DNA thermal cycler called Mastercycler Personal (Eppendorf, Germany) with the following program: 95℃ for 2 min and 30 cycles of 95℃ for 30 seconds, annealing at 55℃ or 60℃ for 1 min for p28 and dsb, respectively; and 72℃ for 2 min, followed by a final extension at 72℃ for 5 min. The amplified products were separated on a 1.5% agarose gel and visualized by ethidium bromide (Sigma-Aldrich, USA) staining under UV illumination. The amplified dsb and p28 gene products were purified with a QIAquick Gel Extraction Kit (Qiagen, Germany). The DNA was eluted with 10 µL of ultrapure water, quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, USA), and stored at -20℃.
The purified partial sequences of dsb and p28 genes from E. canis Uberlândia were cloned with the pGEM-T Easy Vector System (Promega, USA) following the manufacturer's instructions. Plasmids were isolated from the clones as previously described [9] and sequenced using the DYEnamic ET Dye Terminator Kit (Amersham Pharmacia Biotech, UK) with a MegaBACE 1000 automated sequencer (Amersham Pharmacia Biotech, UK).
All DNA sequences were assembled using the CAP3 Contig Assembly Program [14] with BioEdit Software and then analyzed using BLAST N algorithms [6]. Amino acid sequence prediction was performed using the ExPASy Proteomics Server program [8]. The nucleotide and protein sequence alignments were carried out using ClustalW [31]. Deduction of the antigenicity indices for the protein sequences was performed with LASERGENE Software (DNASTAR, USA) using the Jameson-Wolf antigenicity algorithm [15].
The origins and GenBank (National Center for Biotechnology Information, USA) database accession numbers for the dsb and p28 nucleotide sequences of E. canis strains used for comparison in this study are as follows: two Brazilian strains (Jaboticabal, DQ460716, EF014897 and São Paulo, DQ460715, DQ460713), two North American strains (Oklahoma, AF403710, AF078553 and Jake, CP000107), an African strain (Cameroon, DQ124260), and a Venezuelan strain (VHE, AF165815). The nucleotide sequences we obtained were submitted to GenBank under the accession numbers GU586135 and GU951532 for dsb and p28, respectively.
Forty-eight random serum samples from dogs seen at the Veterinary Hospital of the Federal University of Uberlândia along with random serum samples from 60 dogs from the São Paulo metropolitan area were used to assess antigenicity between E. canis Uberlândia and São Paulo.
The antigens for the IFA were produced by separately culturing the E. canis São Paulo and Uberlândia strains in DH82 cells. In summary, these two strains were propagated in uninfected DH82 cells (provided by Marcelo B. Labruna, University of São Paulo) in Dulbecco's Modified Eagle's medium (Sigma-Aldrich, USA) supplemented with 2.5% heat-inactivated bovine calf serum (Hyclone Laboratories, USA). The culture was incubated at 37℃ in a 5% CO2 and the culture medium was partially (20%) replaced every 2~3 days. Cell cultures were monitored once a week for the presence of ehrlichial morulae by Diff-Quik staining according to the manufacturer's instructions (Laborclin Produtos para Laboratórios, Brazil). The cells were harvested using a cell scraper when the percentage of infected cells in the supernatant was 80~90%. The cells were collected by centrifugation at 4,000 g for 5 min at 4℃, and washed three times in PBS (0.0084 M Na2HPO4, 0.0018 M NaH2PO4, and 0.147 M NaCl; pH 7.2). Next, the cells were diluted in PBS to a concentration of 106 cells/mL.
Ten µL of the suspensions were placed into each well of 12-well Teflon (E.I. DuPont de Nemours & Co., USA)-coated multiwell slides (Cell-Line Associates, USA). The slides were dried overnight at room temperature, stored at -20℃, and used within 1 month of preparation. Serial dilutions of each serum sample from an initial dilution of 1 : 40 (two-fold) to 1 : 81,920 were tested. For the negative control, serum from a healthy dog with no history of ticks that was negative for 16S rRNA was used. For the positive control, serum from a dog with CME confirmed by the presence of morulae in blood smears and PCR analysis of 16S rRNA was used. The slides were incubated at 37℃ and 20 µL of the diluted sera were placed in each well. After an incubation period of 30 min at 37℃, the slides were washed three times in PBS (5 min per wash). Five µL of fluorescein isothiocyanate-conjugated anti-dog IgG (Sigma-Aldrich, USA) was added at a dilution of 1 : 100 in PBS, and the slides were incubated for 30 min at 37℃. After washing with PBS as described above, the slides were air-dried and mounted with Prolong Gold anti-fade solution (Invitrogen, USA) and blindly observed by the same person (Rieck, SE) with a fluorescence Axiovert 200 M microscope (Zeiss, Germany).
Data were analyzed using the GraphPad Prism 4 software package (GraphPad Software, USA). Unweighted kappa-coefficients were calculated to evaluate the agreement among the IFA results and serum samples of dogs from Uberlândia and São Paulo. By convention, K values used were: >0 = no agreement, 0~0.19 = poor agreement, 0.20~0.39 = fair agreement, 0.40~0.59 = moderate agreement, 0.60~0.79 = substantial agreement, and 0.80~1.00 = almost perfect agreement [19]. Endpoint titers of the samples positive according to IFA using antigens from two Brazilian E. canis strains (Uberlândia and São Paulo) were compared by Student's t-test. Differences were considered statistically significant when p < 0.05.
No morulae were observed in the peripheral blood smear from the dog with CME symptoms. However, nested PCR specific for the 16S rRNA gene produced a 365-bp product from the blood sample and from the cell culture of DH82 after 13 days of inoculation with leukocytes isolated from infected dog, which were indicative of E. canis infection. One month later, cytoplasmic inclusions associated with E. canis were detected in infected DH82 cells (Fig. 1). The isolate replicated slowly and achieved a 90~100% infection rate after only 3 months. Infected cells were subsequently cultured several times, and 90~100% infection rates were obtained in 15 days.
The partial dsb and p28 gene sequences from the Uberlândia E. canis strain were aligned with the sequences of other Ehrlichia strains accessible in GenBank. Table 1 shows the identity among these sequences. No polymorphism was observed in the dsb gene sequence (data not shown).
The partial p28 gene sequence of E. canis Uberlândia contained nine polymorphisms compared to E. canis São Paulo, E. canis Jaboticabal, E. canis Jake, E. canis Oklahoma and E. canis VHE (Venezuelan human ehrlichia). Additionally, only three differences relative to E. canis Jaboticabal (Fig. 2) were observed. Overall, specific differences among the Uberlândia strain and almost all E. canis strains evaluated in this study were found at eight amino acid positions (Table 2). Antigenic determinants were predicted based on the linear surface contour profile of the deduced p28 amino acid sequences of the E. canis strains. Polymorphisms between these amino acid sequences resulted in few differences in antigenicity (Fig. 3).
Out of the 48 Uberlândia serum samples, 24 were positive (50%) and 24 were negative (50%). Among the samples from 60 dogs from São Paulo, nine (15%) were positive and 51 (85%) were negative. Only seven (21.2%) of the 33 positive sera showed endpoint titers that were different at more than two sera dilution factors between the two antigens (data not shown). For the samples from Uberlândia, a significant difference between the mean and standard deviation of the first and final antigen titer of the positive samples, using both antigen from Uberlândia as São Paulo, was observed (Table 3).
If the reaction was only classified positive or negative, there was no difference between the antigens regardless of the antigen and origin of the serum (kappa = 1.0), indicating that the IFA is able to identify positive animals regardless of the origin of the antigen applied. However, when the results were evaluated based on the maximum positive dilution (using only positive animals), the kappa index was 0.1291, which was considered a poor agreement between the IFA results with heterologous sera. Using only positive sera of dogs from Uberlândia, the index value fell to 0.0247 (very poor agreement) when applied to sera from dogs of São Paulo, whereas the index value was 0.3750 (fair agreement).
In vitro cultivation of E. canis has been widely performed using canine DH82 cells [1,2,5,17,32,33]. In the present study, a new Uberlândia E. canis strain was observed in infected DH82 cells after the same period of time (30 days after inoculation) as previously reported for E. canis from Spain [5] and São Paulo [1]. Partial sequence of the p28 gene from the Uberlândia E. canis strain differed at some nucleotides compared to published sequences. These findings are in agreement with data from a previous study in which a small number of substitutions in the p28 gene from the São Paulo strain was observed when compared with isolates from Jake, Oklahoma, and Venezuela [1]. These results are expected because while the dsb gene is highly conserved among E. canis strains from different countries and continents [1,2,18,28], the p28 gene that encodes an immunodominant protein may be under immune pressure from different hosts and thus exhibit some variation between strains [12,35]. Furthermore, previous analyses of the TRP36 gene, which encodes immunoreactive proteins associated with functional host-pathogen interactions, have shown that this gene is useful for genotyping E. canis strains based on differences in tandem repeat numbers or sequences, indicating some degree of E. canis genetic diversity in nature [3,13,16,26].
When designating the IFA reactions as only positive or negative, the kappa coefficients were in complete agreement independent of the antigen used or origin of the serum evaluated. This finding demonstrated that the antigenic difference observed for the in silico study was not sufficient to generate false negatives or positives. Complete conservation of the p28 gene among seven E. canis isolates from North America suggests that the E. canis gene may be conserved in this geographic area and could be used for diagnostic purposes [21]. Additionally, the Ehrlichia sp. DNA from Brazilian felids was found to be closely related to other strains isolated in Brazil based on the outer membrane protein 1 (omp-1) multigene family [7]. Similar results were produced in our study. The few genetic alterations in the p28 gene of the Uberlândia E. canis strain did not alter the CME diagnostic findings. The lack of genetic diversity among E. canis isolates may be explained by the fact that this microorganism may face little selective pressure because it is predominantly maintained in a single tick species and canine host [22,35].
In contrast, comparison of the final IFA concentration resulted in the identification of weak or poor agreements according to the kappa coefficient, showing that the two antigens produce different responses. Animals from São Paulo, who probably acquired antibodies against the strain from that region, produced a slightly more robust response to the antigen from the São Paulo strain. Similar results were observed with samples from Uberlândia as shown by the fact that only six animals had an endpoint titer for the Uberlândia strain at least 4-fold higher (≥2 serum dilution factors) than the endpoint titers for the São Paulo strain. In contrast, none of the positive sera from São Paulo differed by at least 4-fold (≥2 serum dilution factors) when comparing the two antigens. Similar results were produced by a study in Israel in which titers of sera from 46% of the dogs examined were higher on the locally prepared slides measured by a commercial IFA kit compared to a Florida strain [17]. The use of an autochthonous antigen versus two different antigens increases the sensitivity of the IFA [4]. Furthermore, similarities were observed in lysates from Brazil, the USA, and Israel, which reacted with homologous and heterologous sera, and produced a homogeneous Western immunoblot pattern [36].
In conclusion, findings from the present study suggest that the E. canis genotypes circulating in dogs from southeastern Brazil appear to be highly conserved. The IFA results indicated that both antigens from Uberlândia and São Paulo could be used to identify antibodies against Brazilian E. canis strains. This is because both antigens are able to diagnose positive and negative animals.
Figures and Tables
Fig. 1
Photomicrograph of an Ehrlichia (E.) canis strain isolated from the leukocytes of a dog in Uberlândia, Brazil. Note the multiple round, large morulae in the DH82 cell cytoplasm (arrows). Diff-Quik staining was performed. Scale bar = 10 µm.
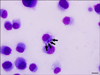
Fig. 2
Clustal alignment of p28 gene sequences from E. canis VHE (Venezuelan human ehrlichia), São Paulo, Jake, Oklahoma, Jaboticabal, and Uberlândia strains. Boxed nucleotides are bases that differ from the Uberlândia strain sequence, and a dash represents a gap or nucleotide that was unidentified.

Fig. 3
Comparison of the antigenic indices for the predicted partial proteins translated from the p28 gene of the following strains: E. canis Uberlândia, São Paulo, Jaboticabal, Jake, Oklahoma, and VHE. Regions with values above zero are possible antigenic determinants. Boxed sequences are potential epitopes that differ from the Uberlândia strain sequence. The scale indicates amino acid positions.

Table 1
Nucleotide sequence identities of dsb and p28 partial gene sequences for the E. canis Uberlândia strain and available E. canis sequences

References
1. Aguiar DM, Hagiwara MK, Labruna MB. In vitro isolation and molecular characterization of an Ehrlichia canis strain from São Paulo, Brazil. Braz J Microbiol. 2008; 39:489–493.

2. Aguiar DM, Saito TB, Hagiwara MK, Machado RZ, Labruna MB. Serological diagnosis of canine monocytic ehrlichiosis with Brazilian antigen of Ehrlichia canis. Cienc Rural. 2007; 37:796–802.
3. Aguiar DM, Zhang X, Melo AL, Pacheco TA, Meneses AM, Zanutto MS, Horta MC, Santarém VA, Camargo LM, McBride JW, Labruna MB. Genetic diversity of Ehrlichia canis in Brazil. Vet Microbiol. 2013; 164:315–321.
4. Aguirre E, Ayllón T, Sainz A, Amasutegui I, Villaescusa A, Rodríquez-Franco F, Tesouro MA. Results from an indirect fluorescent antibody test using three different strains of Ehrlichia canis. Vet J. 2009; 182:301–305.

5. Aguirre E, Sainz A, Dunner S, Amusategui I, López L, Rodríguez-Franco F, Luaces I, Cortés O, Tesouro MA. First isolation and molecular characterization of Ehrlichia canis in Spain. Vet Parasitol. 2004; 125:365–372.

6. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990; 215:403–410.

7. André MR, Adania CH, Machado RZ, Allegretti SM, Felippe PA, Silva KF, Nakaghi AC. Molecular and serologic detection of Ehrlichia spp. in endangered Brazilian wild captive felids. J Wildl Dis. 2010; 46:1017–1023.

8. Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiqer E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaquir K, Redaschi N, Rossier G, Xenarios I, Stockinger H. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012; 40(Web Server issue):W597–W603.

9. Azevedo MA, Felipe MSS, Maranhão AQ, De Souza MT. Técnicas Básicas em Biologia Molecular. 1st ed. Brasília: Editora Universidade de Brasília;2003. p. 49–72.
10. Costa OJ, Batista JA Jr, Silva M, Guimarães MP. Ehrlichia canis infection in dog in Belo Horizonte - Brazil. Arquivos da Escola de Veterinária da UFMG. 1973; 25:199–206.
11. Crocquet-Valdes PA, Thirumalapura NR, Ismail N, Yu X, Saito TB, Stevenson HL, Pietzsch CA, Thomas S, Walker DH. Immunization with Ehrlichia p28 outer membrane proteins confers protection in a mouse model of ehrlichiosis. Clin Vaccine Immunol. 2011; 18:2018–2025.

12. Doyle CK, Labruna MB, Breitschwerdt EB, Tang YW, Corstvet RE, Hegarty BC, Bloch KC, Li P, Walker DH, McBride JW. Detection of medically important Ehrlichia by quantitative multicolor TaqMan real-time polymerase chain reaction of the dsb gene. J Mol Diagn. 2005; 7:504–510.

13. Hsieh YC, Lee CC, Tsang CL, Chung YT. Detection and characterization of four novel genotypes of Ehrlichia canis from dogs. Vet Microbiol. 2010; 146:70–75.

15. Jameson BA, Wolf H. The antigenic index: a novel algorithm for predicting antigenic determinants. Comput Appl Biosci. 1988; 4:181–186.

16. Kamani J, Lee CC, Haruna AM, Chung PJ, Weka PR, Chung YT. First detection and molecular characterization of Ehrlichia canis from dogs in Nigeria. Res Vet Sci. 2013; 94:27–32.

17. Keysary A, Waner T, Rosner M, Warner CK, Dawson JE, Zass R, Biggie KL, Harrus S. The first isolation, in vitro propagation, and genetic characterization of Ehrlichia canis in Israel. Vet Parasitol. 1996; 62:331–340.

18. Labruna MB, McBride JW, Camargo LM, Aguiar DM, Yabsley MJ, Davidson WR, Stromdahl EY, Williamson PC, Stich RW, Long SW, Camargo EP, Walker DH. A preliminary investigation of Ehrlichia species in ticks, humans, dogs and capybaras from Brazil. Vet Parasitol. 2007; 143:189–195.

19. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977; 33:159–174.

20. McBride JW, Ndip LM, Popov VL, Walker DH. Identification and functional analysis of an immunoreactive DsbA-like thio-disulfide oxidoreductase of Ehrlichia spp. Infect Immun. 2002; 70:2700–2703.

21. McBride JW, Yu XJ, Walker DH. Molecular cloning of the gene for a conserved major immunoreactive 28-kilodalton protein of Ehrlichia canis: a potential serodiagnostic antigen. Clin Diagn Lab Immunol. 1999; 6:392–399.

22. McBride JW, Walker DH. Progress and obstacles in vaccine development for the ehrlichioses. Expert Rev Vaccines. 2010; 9:1071–1082.

23. Murphy GL, Ewing SA, Whitworth LC, Fox JC, Kocan AA. A molecular and serological survey of Ehrlichia canis, E. chaffeensis, and E. ewingii in dogs and ticks from Oklahoma. Vet Parasitol. 1998; 79:325–339.

24. Nazari M, Lim SY, Watanabe M, Sharma RSK, Cheng NABY, Watanabe M. Molecular detection of Ehrlichia canis in dogs in Malaysia. PLoS Negl Trop Dis. 2013; 7:e1982.
25. Ohashi N, Zhi N, Zhang Y, Rikihisa Y. Immunodominant major outer membrane proteins of Ehrlichia chaffeensis are encoded by a polymorphic multigene family. Infect Immun. 1998; 66:132–139.

26. Rar V, Golovljova I. Anaplasma, Ehrlichia, and "Candidatus Neoehrlichia" bacteria: pathogenicity, biodiversity, and molecular genetic characteristics, a review. Infect Genet Evol. 2011; 11:1842–1861.

27. Rikihisa Y, Ewing SA, Fox JC. Western immunoblot analysis of Ehrlichia chaffeensis, E. canis, or E. ewingii infections in dogs and humans. J Clin Microbiol. 1994; 32:2107–2112.

28. Romero LE, Meneses AI, Salazar L, Jiménez M, Romero JJ, Aguiar DM, Labruna MB, Dolz G. First isolation and molecular characterization of Ehrlichia canis in Costa Rica, Central America. Res Vet Sci. 2011; 91:95–97.

29. Santos F, Coppede JS, Pereira AL, Oliveira LP, Roberto PG, Benedetti RB, Zucoloto LB, Lucas F, Sobreira L, Marins M. Molecular evaluation of the incidence of Ehrlichia canis, Anaplasma platys and Babesia spp. in dogs from Ribeirão Preto, Brazil. Vet J. 2009; 179:145–148.

30. Thomas S, Thirumalapura NR, Crocquet-Valdes PA, Luxon BA, Walker DH. Structure-based vaccines provide protection in a mouse model of ehrlichiosis. PLoS One. 2011; 6:e27981.

31. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22:4673–4680.

32. Torres HM, Massard CL, de Figueiredo MJ, Ferreira T, Almosny NRP. Isolation and propagation of Ehrlichia canis in DH82 cells as a source of antigens in indirect immunofluorescence test. Revista brasileira de ciência veterinária. 2002; 9:77–82.
33. Unver A, Perez M, Orellana N, Huang H, Rikihisa Y. Molecular and antigenic comparison of Ehrlichia canis isolates from dogs, ticks and human in Venezuela. J Clin Microbiol. 2001; 39:2788–2793.

34. Vieira RF, Biondo AW, Guimarães AM, Dos Santos AP, Dos Santos RP, Dutra LH, Diniz PP, de Morais HA, Messick JB, Labruna MB, Vidotto O. Ehrlichiosis in Brazil. Rev Bras Parasitol Vet. 2011; 20:1–12.

35. Yu XJ, McBride JW, Walker DH. Restriction and expansion of Ehrlichia strain diversity. Vet Parasitol. 2007; 143:337–346.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download