Abstract
This study was conducted to investigate the expression of three genes related to early embryonic development in bovine transgenic cloned embryos. To accomplish this, development of bovine transgenic somatic cell nuclear transfer (SCNT) embryos was compared with non-transgenic embryos. Next, mRNA transcription of three specific genes (DNMT1, Hsp 70.1, and Mash2) related to early embryo development in transgenic SCNT embryos was compared between transgenic and non-transgenic SCNTs, parthenogenetic embryos, and in vitro fertilization (IVF) embryos. Transgenic SCNT embryos showed significantly lower rates of development to the blastocyst stage than non-transgenic ones. To investigate normal gene expression, RNA was extracted from ten blastocysts derived from parthenogenesis, IVF, non-transgenic, and transgenic SCNT embryos and reverse-transcribed to synthesize cDNA. The cDNA was then subjected to PCR amplification and semi-quantified. More DNMT1 mRNA was detected in the transgenic SCNT group than the other three groups. Hsp 70.1 mRNA was detected in the IVF embryos, while lower levels were found in SCNT and parthenogenetic embryos. Mash2 mRNA was present at the highest levels in transgenic SCNT embryos. In conclusion, the higher levels of methylation and lower protein synthesis after heat shock in the transgenic SCNT embryos expected based on our results may cause lower embryonic development.
There has been great progress in generation of bovine somatic cell nuclear transfer (SCNT) embryos using fetal fibroblasts or adult somatic cells as donor cells since the first reports of cloned mammals [7,18,35]. Transgenic cows produced by SCNT have been reported to produce human pharmaceutical proteins in their milk as bioreactors [13]. However, it has consistently been reported that high rate of embryonic, fetal, and neonatal abnormalities in the bovine SNCT, often within the context of large offspring syndrome [7,9,21,34]. For future successful development of transgenic bovine cloned embryos, many factors and their impacts on embryo quality must be considered during the SCNT procedure [5].
In studies of bovine transgenic SCNT, different development rates were shown between transgenic SCNT using transfected cells and control SCNT using non-treated somatic cells [2,3]. No significant differences in developmental competence were observed between transgenic and non-transgenic SCNT in studies conducted by Arat et al. [2,3]. However, bovine transgenic SCNT embryos showed significantly lower development competence than non-transgenic SCNT in a study conducted by Zakhartchenko et al. [40]. During SCNT, reprogramming of injected donor cells is essential for further embryonic development after SCNT. This nuclear reprogramming includes structural remodeling and is related to fundamental changes in genome activity [16]. In this process, both proteins are exchanged between recipient cytoplasm and donor nuclei, and rapid swelling of transplanted nuclei occurs [12]. Complete reprogramming of nuclear fine structure was shown in NT-derived bovine embryos through previous ultrastructural studies [19,20,30].
Normal expression of genes related to embryonic development is needed when reprogramming donor cells after SCNT. To date, the molecular mechanisms related to nuclear reprogramming in SCNT embryos have remained largely unknown [23,39]. Genetic imprinting [31] and epigenetic DNA modifications, including DNA methylation [26] and changes in chromatic configuration [22], likely play a role in reprogramming of the donor nucleus. Additionally, altered transcript levels were found for several genes related with embryonic devleopment in bovine NT embryos [10]. Thus, determination of the expression patterns of essential genes in donor nuclear reprogramming is needed for efficient production of bovine SCNT embryos. This information can be further applied in transgenic SCNT as well.
In the present study, developmental competence of transgenic SCNT embryos was compared with that of non-transgenic SCNT embryos. In addition, the expression of genes related to embryo development were compared in parthenogenetic, in vitro fertilization (IVF), non-transgenic SCNT, and transgenic SCNT embryos to investigate the reasons for different developmental competence between groups. Transcription of DNA methyltransferase (DNMT), heat shock protein (Hsp) 70.1, and mammalian achaete-scute homologue (Mash2) were evaluated. The DNMT1 gene is related to DNA methylation [38], Hsp 70.1 is expressed under the stress of temperature changes, [17] and Mash2 transcription is related to early embryonic differentiation and trophoblastic function [1]. The identification of differential expression in these genes that are frequently abnormally expressed in transgenic NT embryos will provide valuable information regarding the cause of abnormality in transgenic SCNT embryos and facilitate development of methods to increase transgenic SCNT embryo production efficiency.
Generation of human prourokinase gene-transfected bovine cells was performed as previously described [6,8]. Briefly, we collected cumulus cells from cumulus oocyte complexes (COCs) by follicle aspiration guided with ultrasonography [24]. We washed cumulus cells once by centrifugation and then seeded them into 100 mm Falcon plastic culture dishes. We then cultured seeded cells for 3~4 days in DMEM supplemented with 10% fetal bovine serum, 1 mM sodium pyruvate, 1% non-essential amino acids, and 10 µg/mL penicillin streptomycin solution, after which we removed the explants. Dissociated cells were sequentially plated in new Petri dishes containing the same culture medium, after which cumulus cells were transfected through lipid-mediated gene transfer. First, the pGFP-proU plasmid containing the enhanced green fluorescent protein reporter gene driven by the cytomegalovirus promoter and human prourokinase gene driven by the bovine beta casein promoter was delivered into the cells using FuGene6 (Roche Molecular Biochemicals) according to the manufacturer's instructions. Two days later, cells were examined under ultraviolet light to evaluate transfection. Transfected cells were subsequently collected by trypsinization of the monolayer and centrifuged, after which cell pellets were resuspended in PBS supplemented with 0.5% FBS until SCNT.
Bovine ovaries were collected at a local slaughterhouse and transported to the laboratory in 0.9% (v/v) NaCl solution at 30℃ to 35℃ as previously described [5]. Follicular fluid and COCs from follicles 2 to 8 mm in diameter were aspirated using an 18-gauge needle attached to a 10 mL disposable syringe. COCs with evenly granulated cytoplasm enclosed by compact cumulus cells of more than three layers were selected, washed three times in HEPES-buffered tissue culture medium-199 supplemented with 10% FBS, 0.005 AU/mL FSH (Antrin; Teikoku, Japan), and 1 µg/mL estradiol (Sigma, USA) and then incubated at 39℃ in a humidified atmosphere of 5% CO2 and 95% air for 18 h.
Four types of embryos were produced in this experiment as previous described [6].
After 24 h of maturation, oocytes were denuded by mouth pipetting, after which mature oocytes were selected based on the presence of the first polar body under a stereomicroscope. After 4 h of culture, mature oocytes were activated by incubation in handling medium containing 5 µm ionomycin for 4 min. Embryos were then extensively washed in TCM199-washing medium for 5 min before culture for 4 h in 2 mM 6-DMAP (Sigma) in modified synthetic oviductal fluid (mSOF) for postactivation. Culture of parthenogenetic embryos was performed in 25 µL drops of mSOF overlaid with mineral oil at 39℃ for 7 days in a humidified atmosphere of 5% CO2, 5% O2, and 90% N2.
TALP medium containing 3 or 6 mg/ml BSA, as described by Fukui [14], was used for oocyte washing, sperm penetration, and IVF. After 22 h of maturation, mature oocytes were rinsed three times in 2~3 mL washing-TALP medium and then rinsed once in IVF-TALP. Five to seven oocytes were transferred into a 43 µL droplet of IVF-TALP under mineral oil. Frozen-thawed sperm were subjected to a swim-up procedure for 1 h in capacitation-TALP to recover motile spermatozoa. The supernatant was then centrifuged at 500 × g for 6 min and a 5 µL aliquot of the sperm pellet was added to the IVF-TALP (approximately 1 × 106/mL) droplets containing the oocytes. Fertilization was allowed to take place at 39℃ in a humidified atmosphere of 5% CO2 in air for 18~20 h. After culture, fertilized oocytes were denuded by gentle pipetting in a TCM-washing medium. Embryos were then cultured in 25 µL drops of mSOF overlaid with mineral oil at 39℃ for 7 days in a humidified atmosphere of 5% CO2, 5% O2, and 90% N2.
Production of non-transgenic SCNT embryos was performed as described by Cho et al. [7]. Briefly, after 18 h of maturation, oocytes were denuded and mature oocytes with a first polar body were selected for enucleation, which was conducted by the squeezing method. Following enucleation, cumulus donor cells were injected into enucleated oocytes. These reconstructed embryos were then fused electrically and cultured for reprogramming followed by activation using a 5 µM ionomycin. At 4 h postactivation, embryos were cultured in 25 µL drops of mSOF overlaid with mineral oil at 39℃ for 7 days in a humidified atmosphere of 5% CO2, 5% O2 and N2 in an air atmosphere.
Total RNA was extracted by the acid phenol-guanidinium thiocyanate-chloroform extraction method [28]. Sets of 10 parthenogenetic-, IVF-, non-transgenic SCNT-, and transgenic SCNT-blastocysts that had been cultured for 7 days were added to 200 µL guanidine isothiocyanate solution (Invitrogen, USA) supplemented with 1% (v/v) β-mercaptoethanol (Sigma) and stored at -80℃ deep freezer. Samples were gently mixed with the same volume of phenol chloroform isothiocyanate (PCI) medium for RNA analysis. After centrifugation of this mixed solution at 18,890 × g and 4℃ for 10 min, 140 µL of 100% isopropanol (Sigma) and 1 µL glycogen (Invitrogen) were added and stored at -20℃ overnight. Following centrifugation of this mixture at 18,890 × g and 4℃ for 40 min, supernatant was removed and RNA was washed with 700 µL of 70% (v/v) ethanol. After removal of the ethanol, total RNA was eluted by adding 15 µL RNAase-free water. First-strand cDNA for reverse transcription (RT) was then synthesized using the first-strand cDNA synthesis kit (Amersham Phamarcia Biotech) according to the manufacturer's protocols. Briefly, eluted RNA was heated to 65℃ for 10 min for mRNA denaturation and then immediately transferred to ice before the addition of RT reagents, including 11 µL bulk first-strand cDNA reaction mix, 1 µL of DTT solution, and 1 µL of first-strand cDNA primer (Not1-d(T)18). After mixing and centrifugation, the reaction mixture was incubated at 37℃ for 1 h.
Polymerase chain reaction (PCR) was performed using first strand cDNA synthesized from embryos generated by parthenogenetic activation, IVF, non-transgenic SCNT, and transgenic SCNT in a final volume of 50 µL composed of 36.5 µL distilled water, 5 µL 10× PCR Buffer (Qiagen, Germany), 2 µL 200 µM each Advantage UltraPure dNTP Mix (Clontech Laboratories USA), and 1 µL of 50 pmoles/µL sequence-specific primers. The primer sequences of three genes and β-actin as housekeeping gene were summarized in Table 1 [37]. The PCR program consist of an initial step of 95℃ for 15 min and various cycle numbers of denaturing (95℃ for 50 sec), annealing (each different temperature for 50 sec), extension (72℃ for 60 sec), and final extension for 15 min at 72℃. As negative controls, tubes were prepared without RNA or reverse transcriptase. The RT-PCR products were subjected to electrophoresis on 2% agarose gel in 1× TAE buffer (45 mM Tris-acetate, 1 mM EDTA, pH 8.3) containing 0.2 µg/mL ethidium bromide. The image of each gel was recorded using a Gel Doc 2000 Gel Documentation System (BioRad Laboratories, USA) with Quantity One program.
Multiple comparisons were implemented using Generalized Linear Models in the SAS 8.12. program (SAS Institute, USA). When significant main effects in each experimental parameter were detected, subsequent comparisons were made by the least squares method. Differences among treatments were considered statistically significant at p < 0.05. Semi-quantitative gene expression in the four types of embryos was analyzed by one-way ANOVA using SigmaStat 2.0 (Systat Software, USA).
The developmental competence of non-transgenic and transgenic SCNT embryos is summarized in Table 2. There were no significant differences in fusion rates between non-transgenic SCNT embryos and transgenic SCNT embryos (69.0% vs. 78.4%). However, there were significant differences in cleavage rates and development rates to blastocyst between SCNT groups (61.3% vs. 73.4%; 43.3% vs. 28.7%; p < 0.05). Transgenic SCNT embryos showed higher cleavage rates, but significantly lower development rates (p < 0.05).
Representative gel photos of transcripts of each 3 gene with relative comparison with β-actin as housekeeping gene in parthenogenetic, IVF, control NT, and transgenic NT blastocysts are shown in Figs. 1,2,3. To analyze quantitative differential expression of the three selected genes related to embryo development, expression values of each band were divided into ones of β-actin (control gene). DNMT1 mRNA was detected in all groups, and was relatively more abundant in the transgenic SCNT group than the other three groups (Fig. 1). Hsp70.1 mRNA was not detectable in the parthenogenetic and NT embryos, whereas Hsp70.1 expression was observed in IVF embryos (Fig. 2). Furthermore, in IVF embryos, the relative abundance of Hsp70.1 transcripts was higher than that of the other three groups. Mash2 mRNA was detected in both NT blastocysts, but was less abundant in parthenogenetic and IVF embryos (Fig. 3). Transcripts of transgenic SCNT embryos were generally higher than those of non-transgenic SCNT embryos.
Developmental competence of transgenic SCNT embryos is normally significantly lower than that of control SCNT embryos, which was corroborated by the results of the present study. To determine the genetic causes of low production rates by SCNT techniques, transcription levels of the three genes were evaluated. DNMT1, Hsp70.1, and Mash2 relative expression was examined in bovine preimplantation embryos derived from parthenogenetic activation, IVF, non-transgenic SCNT, and transgenic SCNT procedures. Transcription levels of these genes that have important functions during pre- and post-implantation development can be used as markers for embryonic viability.
Comparison of developmental rates between transfected donor cells and non-transfected donor cells showed that transfection of donor cells significantly negatively affects developmental rate. In a study conducted by Arat [2,3], there were no differences in development rates of either SCNT embryos. However, in our study, development rates to blastocyst were significantly decreased in the transgenic SCNT embryos (43.3% vs. 28.7%). This discrepancy may have been caused by a difference in the length of the gene that was inserted. Arat used a relatively short transfected gene (4651 bp; pEGFP-N1) than we employed (14200 bp). Additionally, in our pbeta2.1-proUK vector, the bovine β-casein promoter (3200 bp) and the GFP gene (718 bp) were inserted into the pcDNA vector along with our gene of interest. Therefore, prior to transfection of this product into a somatic cell, the donor cell may have been damaged due to a deleterious effect of those gene products. Additionally, exposure to UV light for selection of transfected cells may have influenced the developmental potential of transgenic NT embryos. Smith [29] showed that UV irradiation to the bovine in vitro matured oocytes for more than 30 sec caused alterations to intracellular components and membrane. Bell et al. [4] also demonstrated that UV irradiation decreases protein synthesis in oocytes.
The successful reprogramming of a donor cell in the SCNT by cytoplasmic factors would result with the similar gene expression profile as that seen in the IVF embryos [37]. In preimplantation development of SCNT embryos, this successful reprogramming could be examined by transcription onset of these important genes that is only required embryonic development with switch off in the differentiated donor cells. Transcription of three genes in blastocysts derived from parthenogenetic, IVF, non-transgenic SCNT, and transgenic SCNT differed.
In the mammalian embryonic development, DNA methylation is essential for regulation of transcription and it is involved in X chromosome inactivation, imprinting, cell differentiation, and gene silencing [33]. DNMT1 is the most abundant type of DNA methyltransferase, and is essential in the copying methylation patterns following DNA synthesis [27]. When compared to IVF control blastocysts, the relative amount of DNMT 1 transcripts was significantly decreased in SCNT embryos, regardless of whether normal or transfected donor cells were used. It could be concluded that abnormally high DNMT1 transcript levels in transgenic SCNT embryos was related with methylation problem after each cleavage from one cell embryo to the blastocyst stage. If this methylation error is persistent to term, this could be associated with the offspring abnormalities by epigenetic changes in the embryonic development after non-transgenic SCNT and transgenic SCNT [25,39]. Hsp 70.1 was reported as one of essential protein is expressed after environmental stress in bovine preimplantation embryos, and was detected in IVF blastocysts in several reports [36,37]. However, in the present study, Hsp70.1 was not detected in either NT embryos. No detection in these embryos could be associated with inability for appropriate response to the environmental stress in the early embryonic development and contribute extremely lower development rates to term. In this study, Mash2 gene was highly expressed in both NT embryos, while it was shown lower transcripts in the IVF and parthenogenetic embryos. The Mash2 gene is highly expressed in the trophoblast cells that is responsible for fetal membrane development, but is seen at low levels in the embryo proper for fetus [15]. The results of this study revealed relatively high Mash2 transcripts in transgenic SCNT embryos, suggesting that excessive proliferation of trophoblasts is related to large offspring syndrome that is usually seen in the cloned cow.
In conclusion, our results showed relatively lower development rates in transgenic SCNT embryos compared to non-transgenic SCNT embryos. By comparing the mRNA expression of three critical embryonic developmental genes in blastocysts derived from parthenogenetic, IVF, non-transgenic SCNT, and transgenic SCNT, we demonstrated variation in the expression pattern of these important genes, particularly between transgenic and non-transgenic SCNT embryos. The results of this study showed that mRNA transcripts levels can be useful indicator to elucidate the effects of SCNT at the molecular level, and this line of study will aid future transgenic work by increasing the proportion of viable calves without abnormal development such as large offspring syndrome.
Figures and Tables
Fig. 1
Representative gel photograph of semi-quantitative RT-PCR of DNA methyltransferase (DNMT1) transcripts in transgenic NT, control NT, in vitro firtilization (IVF), and parthenogenetic bovine blastocysts. (A) Band of DNMT1 mRNA transcripts. (B) Band of β-actin transcripts as a control. (C) Relative abundance of DNMT1 mRNA transcripts. Expected PCR products of DNMT1 gene (310 bp) in transgenic NT (Lane 1), control NT (Lane 2), IVF (Lane 3), and parthenogenetic (Lane 4) blastocysts, respectively, are indicated (arrow). RT-PCR products were analyzed on 2.0% agarose gel.

Fig. 2
Representative gel photograph of semi-quantitative RT-PCR of heat shock protein 70.1 (Hsp 70.1) transcripts in transgenic NT, control NT, IVF, and parthenogenetic bovine blastocysts. (A) Band of Hsp mRNA transcripts. (B) Band of β-actin transcripts as a control. (C) Relative abundance of Hsp mRNA transcripts. Expected PCR products of Hsp 70.1 gene (488 bp) in transgenic NT (Lane 1), control NT (Lane 2), IVF (Lane 3), and parthenogenetic (lane 4) blastocysts, respectively, are indicated (arrow). RT-PCR products were analyzed on 2.0% agarose gel.
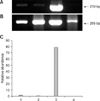
Fig. 3
Representative gel photograph of a semi-quantitative RT-PCR of mammalian achaete-scute homologue (Mash2) transcripts in transgenic NT, control NT, IVF, and parthenogenetic bovine blastocysts. (A) Band of Mash2 mRNA transcripts. (B) Band of β-actin transcripts as a control. (C) Relative abundance of Mash2 mRNA transcripts. Expected PCR products of Mash2 gene (210 bp) in transgenic NT (Lane 1), control NT (Lane 2), IVF (Lane 3), and parthenogenetic (Lane 4) blastocysts was indicated (arrow), respectively. RT-PCR products were analyzed on a 2.0% agarose gel.

Table 2
Development rates of nuclear transfer embryos derived from non-transfected and transfected cumulus cells

Model effects of transfection on the number of embryos fused, cleaved, and developed to blastocysts, which were indicated as p values, were 0.1238, 0.0001, and 0.0001, respectively. aPercentage of the number of embryos injected. bPercentage of the number of embryos fused. cdWithin a parameter in the same treatment, values with different superscripts differed significantly, p < 0.05.
Acknowledgments
This research was supported by the Bio-industry Technology Development Program (111078-3), the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries, Ministry of Agriculture, Food and Rural Affairs, Korea. In addition, we thank Joseph Cacciopo for his valuable editing of this manuscript.
References
1. Alders M, Hodges M, Hadjantonakis AK, Postmus J, van Wijk I, Bliek J, de Meulemeester M, Westerveld A, Guillemot F, Oudejans C, Little P, Mannens M. The human Achaete-Scute homologue 2 (ASCL2, HASH2) maps to chromosome 11p15.5, close to IGF2 and is expressed in extravillus trophoblasts. Hum Mol Genet. 1997; 6:859–867.

2. Arat S, Gibbons J, Rzucidlo SJ, Respess DS, Tumlin M, Stice SL. In vitro development of bovine nuclear transfer embryos from transgenic clonal lines of adult and fetal fibroblast cells of the same genotype. Biol Reprod. 2002; 66:1768–1774.

3. Arat S, Rzucidlo SJ, Gibbons J, Miyoshi K, Stice SL. Production of transgenic bovine embryos by transfer of transfected granulosa cells into enucleated oocytes. Mol Reprod Dev. 2001; 60:20–26.

4. Bell JC, Smith LC, Rumpf R, Goff AK. Effect of enucleation on protein synthesis during maturation of bovine oocytes in vitro. Reprod Fertil Dev. 1997; 9:603–608.

5. Cho J, Bhuiyan MM, Shin S, Park E, Jang G, Kang S, Lee B, Hwang W. Development potential of transgenic somatic cell nuclear transfer embryos according to various factors of donor cell. J Vet Med Sci. 2004; 66:1567–1573.

6. Cho JK, Bhuiyan MMU, Jang G, Park ES, Kang SK, Lee BC, Hwang WS. Production of bovine transgenic cloned embryos using prourokinase-transfected somatic cells: effect of expression level of reporter gene. Korean J Embryo Transf. 2002; 17:101–108.
7. Cho JK, Lee BC, Park JI, Lim JM, Shin SJ, Kim KY, Lee BD, Hwang WS. Development of bovine oocytes reconstructed with different donor somatic cells with or without serum starvation. Theriogenology. 2002; 57:1819–1828.

8. Cho J, Song B, Yong H, Lee D, Koo D, Lee K, Shin S. Differential gene expression in the bovine transgenic nuclear transfer embryos. J Vet Clin. 2007; 24:295–299.
9. Cibelli JB, Stice SL, Golueke PJ, Kane JJ, Jerry J, Blackwell C, Ponce de León FA, Robl JM. Cloned transgenic calves produced from nonquiescent fetal fibroblasts. Science. 1998; 280:1256–1258.

10. Daniels R, Hall V, Trounson AO. Analysis of gene transcription in bovine nuclear transfer embryos reconstructed with granulosa cell nuclei. Biol Reprod. 2000; 63:1034–1040.

11. Dean W, Bowden L, Aitchison A, Klose J, Moore T, Meneses JJ, Reik W, Feil R. Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: association with aberrant phenotypes. Development. 1998; 125:2273–2282.

12. Diberardino MA, Hoffner NJ, Matilsky MB. Methods for studying nucleocytoplasmic exchange of nonhistone proteins in embryos. Methods Cell Biol. 1977; 16:141–165.
13. Echelard Y, Williams JL, Destrempes MM, Koster JA, Overton SA, Pollock DP, Rapiejko KT, Behboodi E, Masiello NC, Gavin WG, Pommer J, Van Patten SM, Faber DC, Cibelli JB, Meade HM. Production of recombinant albumin by a herd of cloned transgenic cattle. Transgenic Res. 2009; 18:361–376.

14. Fukui Y. Effect of follicle cells on the acrosome reaction, fertilization, and developmental competence of bovine oocytes matured in vitro. Mol Reprod Dev. 1990; 26:40–46.

15. Guillemot F, Caspary T, Tilghman SM, Copeland NG, Gilbert DJ, Jenkins NA, Anderson DJ, Joyner AL, Rossant J, Nagy A. Genomic imprinting of Mash2, a mouse gene required for trophoblast development. Nat Genet. 1995; 9:235–242.

17. Gutierrez JA, Guerriero V Jr. Chemical modifications of a recombinant bovine stress-inducible 70 kDa heat-shock protein (Hsp70) mimics Hsp70 isoforms from tissues. Biochem J. 1995; 305:197–203.

18. Hirao Y, Naruse K, Kaneda M, Somfai T, Iga K, Shimizu M, Akagi S, Cao F, Kono T, Nagai T, Takenouchi N. Production of fertile offspring from oocytes grown in vitro by nuclear transfer in cattle. Biol Reprod. 2013; 89:57.
19. Kanka J, Fulka J Jr, Fulka J, Petr J. Nuclear transplantation in bovine embryo: fine structural and autoradiographic studies. Mol Reprod Dev. 1991; 29:110–116.

20. Kanka J, Smith SD, Soloy E, Holm P, Callesen H. Nucleolar ultrastructure in bovine nuclear transfer embryos. Mol Reprod Dev. 1999; 52:253–263.

21. Kato Y, Tani T, Sotomaru Y, Kurokawa K, Kato J, Doguchi H, Yasue H, Tsunoda Y. Eight calves cloned from somatic cells of a single adult. Science. 1998; 282:2095–2098.

22. Koide T, Ainscough J, Wijgerde M, Surani MA. Comparative analysis of Igf-2/H19 imprinted domain: identification of a highly conserved intergenic DNase I hypersensitive region. Genomics. 1994; 24:1–8.

24. Lee BC, Yoon KY, Kim JT, Lee KN, Roh SH, Shin TY, Park JY, Kim NR, Joo SC, Baek NY, Lee ES, Lim JM, Lee WK, Hwang WS. Transvaginal ultrasound-guided ovum pick-up (OPU) in cattle. 2. First OPU-IVF derived calves born from pregnant cow in Korea. Korean J Embryo Transf. 1998; 13:77–86.
25. Niemann H, Wrenzycki C. Alterations of expression of developmentally important genes in preimplantation bovine embryos by in vitro culture conditions: implications for subsequent development. Theriogenology. 2000; 53:21–34.

26. Razin A, Shemer R. DNA methylation in early development. Hum Mol Genet. 1995; 4:Spec No. 1751–1755.

27. Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999; 27:2291–2298.

28. Sambrook J, Russell DW. Extraction, purification, and analysis of mRNA from eukaryotic cells. Molecular Cloning: A Laboratory Manual. 3rd ed. New York: Cold Spring Harbor Laboratory Press;2001. p. 7.1–7.88.
29. Smith LC. Membrane and intracellular effects of ultraviolet irradiation with Hoechst 33342 on bovine secondary oocytes matured in vitro. J Reprod Fertil. 1993; 99:39–44.

30. Smith SD, Soloy E, Kanka J, Holm P, Callesen H. Influence of recipient cytoplasm cell stage on transcription in bovine nucleus transfer embryos. Mol Reprod Dev. 1996; 45:444–450.

31. Surani MA, Kothary R, Allen ND, Singh PB, Fundele R, Ferguson-Smith AC, Barton SC. Genome imprinting and development in the mouse. Dev Suppl. 1990; 89–98.

32. Tanaka H. Influence of the timing of blastomere isolation after the removal of nocodazole in bovine nuclear transfer. Theriogenology. 1999; 51:1225–1237.

33. Weissbach A, Ward C, Bolden A. Eukaryotic DNA methylation and gene expression. Curr Top Cell Regul. 1989; 30:1–21.

34. Wells DN, Misica PM, Tervit HR. Production of cloned calves following nuclear transfer with cultured adult mural granulosa cells. Biol Reprod. 1999; 60:996–1005.

35. Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997; 385:810–813.

36. Wrenzycki C, Herrmann D, Carnwath JW, Niemann H. Alterations in the relative abundance of gene transcripts in preimplantation bovine embryos cultured in medium supplemented with either serum or PVA. Mol Reprod Dev. 1999; 53:8–18.

37. Wrenzycki C, Wells D, Herrmann D, Miller A, Oliver J, Tervit R, Niemann H. Nuclear transfer protocol affects messenger RNA expression patterns in cloned bovine blastocysts. Biol Reprod. 2001; 65:309–317.

38. Yen RW, Vertino PM, Nelkin BD, Yu JJ, El-Deiry W, Cumaraswamy A, Lennon GG, Trask BJ, Celano P, Baylin SB. Isolation and characterization of the cDNA encoding human DNA methyltransferase. Nucleic Acids Res. 1992; 20:2287–2291.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download