Abstract
Activation of the innate immune system requires recognition of pathogen-associated molecular patterns, such as NOD-like receptors. The NOD-like receptor protein 3 (NLRP3) inflammasome is involved in induction of the pro-inflammatory cytokine, IL-1β, and subsequent inflammatory responses. NLRP3 inflammasome plays important roles in the inflammatory and innate immune responses associated with autoimmune/inflammatory syndrome. However, analysis of the tissue distribution and expression profiles in BALB/c mice is still incomplete. In this study, we investigated the tissue distribution and expression pattern of NLRP3 in BALB/c mice to further elucidate its function in innate immunity in this commonly used laboratory animal model. NLRP3 mRNA expression levels and tissue distribution of the protein were investigated by real-time quantitative PCR and immunohistochemical analyses, respectively. NLRP3 mRNA expression was higher in the kidney and inguinal lymph nodes than in other tissues. Cytoplasmic expression of NLRP3 was detected in the epithelial reticular cells of the spleen and thymus, lymphocytes in the inguinal lymph nodes, cardiac muscle cells, cerebral cortex neurons, alveolar macrophages, renal tubule cells and liver sinusoidal endothelial cells. The results of this study will assist investigators in interpreting site-specific functions and roles of NLRP3 in inflammatory responses.
Inflammation occurs as part of the immune response to infection, resulting in tissue damage. Inflammation is characterized by infiltration of cells of the innate immune system during the acute phase and of T lymphocytes and plasma cells during chronic inflammation [5,7,15]. The innate immune system plays a crucial role in inflammation and regulates the generation of long-lived adaptive immune responses [5,7]. The danger signals presented by invading microorganisms are recognized by pathogen-associated molecular patterns, such as Toll-like receptors, RIG-I-like receptors and NOD-like receptors (NLRs), leading to activation of the innate immune system [7,9]. In the immune response to infection, NLRs induce the production of important soluble pro-inflammatory mediators such as IL-1β, which stimulate subsequent cell apoptosis [5,7,9,15,29]. The NLR, NOD-like receptor protein 3 (NLRP3), which is also known as NACHT, LRR and PYD domains-containing protein 3 (NALP3), is characterized by a central intermediary nucleotide binding domain (NACHT domain), a pyrindomain (PYD) and the leucine-rich repeat (LRR) receptor domain, and similar structures are observed among other NLRs members [9,12,20]. NLRP3 interacts with cardinal, apoptosis-associated speck-like protein containing a caspase-recruitment domain (ASC) and two caspase-1 molecules to form the NLRP3 inflammasome [1,19]. Recent research has shown that the NLRP3 inflammasome plays important roles in the inflammatory and innate immune responses associated with autoimmune/inflammatory syndrome [4,30], as well as infectious agents such as prions [28], HIV-1 [25,26], hepatitis C virus [3], influenza virus [10,21,27] and measles virus [13]. The NLRP3 inflammasome induces the uncontrolled production of IL-1β, which is harmful to the host and initiates inflammatory and immune processes associated with disease pathogenesis. NLRP3 functions via interaction with the ASC domain to activate caspase-1. Studies have shown that adenosine triphosphate (ATP), nigericin, maitotoxin, Staphylococcus aureus and Listeria monocytogenes [17], RNA [11] and uric acid crystals [18] can stimulate the induction of NLRP3 expression. NLRP3 is expressed in immune cells and chondrocytes [6], granulocytes and non-keratinizing epithelial cells [14] in humans. However, comprehensive analysis of the tissue distribution and expression profiles is required to fully elucidate the site-specific functions of NLRP3 in inflammatory responses and innate immunity.
BALB/c mice are one of the most important laboratory animal species and are widely used for investigation of the mechanisms of infection and immunity in the pathogenesis of many human and animal diseases [8,22,24]. To date, details regarding the cell- and tissue-specific expression of NLRP3 are still conspicuously lacking in this model animal. The present study was conducted to clarify NLRP3 mRNA and protein expression and distribution in a variety of BALB/c mice tissues. Such information will assist investigators in interpreting the site-specific functions of NLRP3 in inflammatory responses.
Thirty-day old SPF BALB/c mice (n = 5) were obtained from the Guangdong Experimental Animal Center, Guangzhou, China. Mice were anesthetized by carbon dioxide and sacrificed for tissue sampling (lung, heart, liver, spleen, kidney and inguinal lymph nodes). Each sample was separated into two parts, one that was immediately frozen in liquid nitrogen for 2 h and then stored at -86℃ until RNA extraction was performed, and another that was fixed in 10% neutralized buffered formalin, dehydrated, embedded in paraffin wax, and sectioned (thickness, 4 µm). All animal experiments were approved by the Institutional Animal Care and Use Committee at South China Agricultural University (Certification No. CNAS BL0011).
Total RNA was isolated from 100 mg of selected tissue samples homogenized with RNA-Solv Reagent (Omega Bio-Tech, Canada) in accordance with the manufacturer's instructions. The RNA preparations were treated with RNase-free DNase I to remove possible contaminating DNA and stored at -86℃. The RNA from each of the tissue samples was reverse transcribed to cDNA using an M-MLV Reverse Transcriptase Kit (Promega, USA) according to the manufacturer's guidelines.
The mRNA expression level of NLRP3 was determined by real-time quantitative PCR (qPCR) amplification of cDNA generated from mouse tissues using specific primers. Expression levels were normalized to the mRNA expression levels of the endogenous housekeeping gene, β-actin. Real-time quantitative PCR was performed as previously described, with an annealing temperature of 54℃ [23]. The following primers were used for qPCR analysis: NLRP3, 5'-ACCAGCCAGAGTGGAATGA-3' (forward) and 5'-GCGTGTAGCGACTGTTGAG-3' (reverse); β-actin, 5'-CATCCGTAAAGACCTCTATGCC AAC-3' (forward) and 5'-ATGGAGCCACCGATCCA CA-3' (reverse). Data are expressed as the mean ± SD from three separate experiments.
Immunohistochemical analysis was performed as previously described [23], with the following modifications: NLRP3 protein expression was detected using polyclonal goat anti-NLRP3 specific antibody (ab4207; Abcam, UK) as the primary detection reagent (diluted 1 : 200) and HRP-conjugated rabbit anti-goat IgG antibody (Cell Signaling Technology, USA) as the secondary detection reagent (diluted 1 : 4,000). The reaction was visualized using a DAB (3,3'-diaminobenzidine-tetrahydrochloride) Peroxidase Substrate Kit (Promega), with incubation for 5 min at ambient temperature.
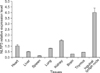
Melting-curve profile analysis confirmed the specificity of primers for PCR amplification of NLRP3 and β-actin fragments. The amplified target gene fragments of 234 and 171 bp, respectively, were evaluated by agarose gel electrophoresis and were in accordance with expectations. The sequences of these two target gene fragments showed 100% homology with the original sequences. NLRP3 mRNA was expressed in all BALB/c mice tissues examined (Fig. 1). The expression of NLRP3 in kidney and lymph nodes was higher than that in other tissues.
Immunohistochemical analysis revealed tissue-specific variation in NLRP3 protein expression, but expression was detected exclusively in the cytoplasm at the cellular level. Almost all splenocytes were negative, and the endothelial cells exhibited weak to medium positive staining in the spleen (Fig. 2A). Similarly, only the endothelial cells of the thymus exhibited weak to medium positive staining (Fig. 2B). Strong positive staining was detected in the lymphocytes of the inguinal lymph nodes (Fig. 2C). In the cerebral cortex of the brain, neurons showed medium to strong positive staining, while only weak staining was detected in some areas of the matrix (Fig. 2D). The type I alveolar epithelial cells of the pulmonary alveoli and alveolar macrophages were moderately to strongly positive for NLRP3 protein expression in the lung (Fig. 2E). In the liver, only a small population of liver cells exhibited medium to strong positive staining, and the liver sinusoidal endothelial cells were strongly positive (Fig. 2F). In the kidney, the cells in the glomerulus and Bowman's capsules were negative, with only the cells of the renal tubules exhibiting strongly positive staining (Fig. 2G). Cardiac muscle exhibited moderate staining, while that of the connective tissues was weakly positive (Fig. 2H).
Although the expression profile and tissue distribution of NLRP3 has been determined in humans [6,14], this information has not yet been made available for BALB/c mice, which are routinely used as model animals in the laboratory. The expression profile of NLRP3 in BALB/c mice may help to elucidate its functions in specific tissues and different disease models. In this study, NLRP3 mRNA can be expressed in all the tested tissues of BALB/c mice. Immunohistological analysis revealed both the tissue and cell distribution of NLRP3 protein expression.
At the mRNA level, NLRP3 was expressed at higher levels in the in kidney and inguinal lymph nodes than in the liver, spleen, lung, brain and thymus. These organs are the main sites of pathogenesis in many diseases, such as autoimmune/inflammatory syndrome [4,30] and infections with prions [28] and viruses [3,10,13,21,27]. Under these conditions, altered expression of NLRP3 and IL-1β can be detected. Investigation of the expression profiles and tissue distribution of NLRP3 in animal models, such as BALB/c mice, is fundamental to elucidation of the function of NLRP3 in innate immunity and inflammation.
Immunohistological analysis of NLRP3 expression in inguinal lymph nodes showed highly positive in lymphocytes, while almost no expression detected in the spleen and thymus in BALB/c mice. It has previously been reported that high NLRP3 mRNA levels were detected in neutrophils, macrophages [2], and primary immune cells [14], while NLRP3 protein was only detected in lymphoid tissues by western blot, and not by IHC, using an in-house generated monoclonal antibody. In 2011, Månsson et al. [16] reported that NALP3 mRNA and protein were present in nasal polyps and normal nasal mucosa in upper airways. Currently, there are no detailed tissue distribution data pertaining to NLPR3 in BALB/c mice. In our study, we confirmed NLRP3 mRNA expression in the lymphoid tissues by qPCR, and demonstrated strong expression of NLRP3 protein in the endothelial cells in the spleen and thymus and very high expression in the lymphocytes of the inguinal lymph nodes by IHC analysis of paraffin-embedded sections of BALB/c mice tissues. It can be speculated that these differences are due to species or detection antibody differences. In further contrast to NLRP3 expression in humans, positive NLRP3 staining of the neurons of the cerebral cortex was located in the cytoplasm, while glial cells, which have been reported to secrete IL-1β [31], were negative [14]. The alveolar macrophages in the lung were positive for NLRP3 expression, which leads to uncontrolled IL-1β maturation and subsequent responses with uncontrolled inflammation demonstrated by the most recent influenza virus infection experiment [21]. A very small population of liver cells in BALB/c mice was confirmed to express NLRP3 by repeated detection, and we have obtained new data demonstrating NLRP3 expression in renal tubules and cardiac muscle cells. Furthermore, our immunohistological data demonstrated that positive staining confirmed that NALP3 are cytoplasmic proteins. In all detected tissues, positive staining was cytoplasmic.
In conclusion, our data showing the expression pattern and tissue distribution of NLRP3 in BALB/c mice should help elucidate the role of different tissues in inflammatory responses and provide a basis for further investigations into the function of NLRP3 in different species.
Figures and Tables
References
1. Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL-1β-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004; 20:319–325.

2. Anderson JP, Mueller JL, Rosengren S, Boyle DL, Schaner P, Cannon SB, Goodyear CS, Hoffman HM. Structural, expression, and evolutionary analysis of mouse CIAS1. Gene. 2004; 338:25–34.

3. Burdette D, Haskett A, Presser L, McRae S, Iqbal J, Waris G. Hepatitis C virus activates interleukin-1β via caspase-1-inflammasome complex. J Gen Virol. 2012; 93:235–246.

4. Chang C. The pathogenesis of neonatal autoimmune and autoinflammatory diseases: a comprehensive review. J Autoimmun. 2013; 41:100–110.

5. Eisenbarth SC, Flavell RA. Innate instruction of adaptive immunity revisited: the inflammasome. EMBO Mol Med. 2009; 1:92–98.

6. Feldmann J, Prieur AM, Quartier P, Berquin P, Certain S, Cortis E, Teillac-Hamel D, Fischer A, de Saint Basile G. Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am J Hum Genet. 2002; 71:198–203.

7. Ferrero-Miliani L, Nielsen OH, Andersen PS, Girardin SE. Chronic inflammation: importance of NOD2 and NALP3 in interleukin-1β generation. Clin Exp Immunol. 2007; 147:227–235.

8. Field J, Biondo MA, Murphy K, Alderuccio F, Toh BH. Experimental autoimmune gastritis: mouse models of human organ-specific autoimmune disease. Int Rev Immunol. 2005; 24:93–110.

9. Fritz JH, Girardin SE. How Toll-like receptors and Nod-like receptors contribute to innate immunity in mammals. J Endotoxin Res. 2005; 11:390–394.

10. Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010; 11:404–410.

11. Kanneganti TD, Özören N, Body-Malapel M, Amer A, Park JH, Franchi L, Whitfield J, Barchet W, Colonna M, Vandenabeele P, Bertin J, Coyle A, Grant EP, Akira S, Núñez G. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006; 440:233–236.

12. Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001; 11:725–732.

13. Komune N, Ichinohe T, Ito M, Yanagi Y. Measles virus V protein inhibits NLRP3 inflammasome-mediated interleukin-1β secretion. J Virol. 2011; 85:13019–13026.

14. Kummer JA, Broekhuizen R, Everett H, Agostini L, Kuijk L, Martinon F, van Bruggen R, Tschopp J. Inflammasome components NALP 1 and 3 show distinct but separate expression profiles in human tissues suggesting a site-specific role in the inflammatory response. J Histochem Cytochem. 2007; 55:443–452.

16. Månsson A, Bogefors J, Cervin A, Uddman R, Cardell LO. NOD-like receptors in the human upper airways: a potential role in nasal polyposis. Allergy. 2011; 66:621–628.

17. Mariathasan S, Weiss DS, Newton K, McBride J, O'Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006; 440:228–232.

18. Martinon F, Pétrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006; 440:237–241.

19. Martinon F, Tschopp J. Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell. 2004; 117:561–574.
20. Martinon F, Tschopp J. NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 2005; 26:447–454.

21. McAuley JL, Tate MD, Mackenzie-Kludas CJ, Pinar A, Zeng W, Stutz A, Latz E, Brown LE, Mansell A. Activation of the NLRP3 inflammasome by IAV virulence protein PB1-F2 contributes to severe pathophysiology and disease. PLoS Pathog. 2013; 9:e1003392.

22. Meyer-Schwesinger C, Dehde S, Klug P, Becker JU, Mathey S, Arefi K, Balabanov S, Venz S, Endlich KH, Pekna M, Gessner JE, Thaiss F, Meyer TN. Nephrotic syndrome and subepithelial deposits in a mouse model of immune-mediated anti-podocyte glomerulonephritis. J Immunol. 2011; 187:3218–3229.

23. Ning Z, An Y, Qi W, Wang H, Pan J, Wu X, Liao M. Na+/H+ exchanger 1 gene expression in tissues of yellow chicken. Biochem Genet. 2012; 50:227–234.

24. Perrone LA, Belser JA, Wadford DA, Katz JM, Tumpey TM. Inducible nitric oxide contributes to viral pathogenesis following highly pathogenic influenza virus infection in mice. J Infect Dis. 2013; 207:1576–1584.

25. Pontillo A, Oshiro TM, Girardelli M, Kamada AJ, Crovella S, Duarte AJS. Polymorphisms in inflammasome' genes and susceptibility to HIV-1 infection. J Acquir Immune Defic Syndr. 2012; 59:121–125.

26. Pontillo A, Silva LT, Oshiro TM, Finazzo C, Crovella S, Duarte AJS. HIV-1 induces NALP3-inflammasome expression and interleukin-1β secretion in dendritic cells from healthy individuals but not from HIV-positive patients. AIDS. 2012; 26:11–18.

27. Pothlichet J, Meunier I, Davis BK, Ting JPY, Skamene E, von Messling V, Vidal SM. Type I IFN triggers RIG-I/TLR3/NLRP3-dependent inflammasome activation in influenza A virus infected cells. PLoS Pathog. 2013; 9:e1003256.

28. Shi F, Yang L, Kouadir M, Yang Y, Wang J, Zhou X, Yin X, Zhao D. The NALP3 inflammasome is involved in neurotoxic prion peptide-induced microglial activation. J Neuroinflammation. 2012; 9:73.

29. Ting JPY, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nat Rev Immunol. 2008; 8:372–379.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download