Abstract
Treatment of bovine mastitis caused by Staphylococcus (S.) aureus is becoming very difficult due to the emergence of multidrug-resistant strains. Hence, the search for novel therapeutic alternatives has become of great importance. Consequently, bacteriophages and their endolysins have been identified as potential therapeutic alternatives to antibiotic therapy against S. aureus. In the present study, the gene encoding lysin (LysSA4) in S. aureus phage SA4 was cloned and the nucleotide sequence was determined. Sequence analysis of the recombinant clone revealed a single 802-bp open reading frame encoding a partial protein with a calculated mass of 30 kDa. Results of this analysis also indicated that the LysSA4 sequence shared a high homology with endolysin of the GH15 phage and other reported phages. The LysSA4 gene of the SA4 phage was subsequently expressed in Escherichia coli. Recombinant LysSA4 induced the lysis of host bacteria in a spot inoculation test, indicating that the protein was expressed and functionally active. Furthermore, recombinant lysin was found to have lytic activity, albeit a low level, against mastitogenic Staphylococcus isolates of bovine origin. Data from the current study can be used to develop therapeutic tools for treating diseases caused by drug-resistant S. aureus strains.
Staphylococcus (S.) aureus is a Gram-positive bacterial pathogen that causes numerous types of infections such as soft tissue abscesses, wound infection, endocarditis, osteomyelitis, bacteremia [18], and mastitis, the most economically significant disease of dairy animals [5]. S. aureus is a cause of great concern due to its low response to antibiotics and capability to persist in herds undetected or at sub-clinical levels [10]. The cure rate of S. aureus mastitis with antibiotics has been reported to be between only 9~35% [6]. S. aureus isolates commonly recovered from clinical cases are not only resistant to β-lactam antibiotics but also similar reagents such as aminoglycosides, fluoroquinolones, and new-generation antibiotics like glycopeptides [2]. In addition to these poor responses to antibiotic treatment, the emergence of bacterial pathogens resistant to chemotherapeutic agents is also a major concern [1]. It is now widely accepted that some previously discontinued methodologies may prove to be useful weapons for the global fight against multidrug-resistant bacteria. Bacteriophages are rapidly re-emerging as possible alternatives to antibiotics for treating bacterial infections [17]. Phage therapy has several advantages over antibiotics including potent in vitro and in vivo antibacterial activity against Gram-positive bacteria [4]. This activity is independent from antibiotic sensitivity patterns, has a low chance of inducing resistance, is easily facilitated by genetic manipulation, and has a high margin of safety.
Bacteriophage endolysins represent a virtually untapped resource for creating new classes of therapeutic agents that are readily available to combat the current antimicrobial resistance crisis in veterinary and human medicine [7]. Phage lysins have attracted considerable interest as novel antibacterial agents, and have been used to control a wide variety of bacteria such as Group A Streptococci [23], Bacillus anthracis [18], Streptococcus pneumonia [12], and Enterococcus faecalis [34]. Bacteriophage produced lytic enzymes that are active against staphylococcal cells have been previously reported [8,27]. A phage-associated lysin could rupture dead and living S. aureus cells [33]. In addition, phage lytic enzymes from staphylococcal phages 80a [3], Twort [14], 187 [13], phi11 [22], vB-SauS-phi-IPLA88 [28,29], and K [26] have been described. The in vitro lytic activity of LysK enzyme was found to be effective against several staphylococcal species including methicillin-resistant S. aureus strains [22].
The lysin gene (plyTW) of the virulent S. aureus phage Twort encoding an enzymatically active fragment was over-expressed and shown to act as an N-acetylmuramyl-L-alanine amidase by cleaving staphylococcal peptidoglycan [14]. Loessner et al. [13] cloned, sequenced, and characterized the S. aureus phage 187 endolysin gene ply187 and reported that this gene (1887 bp) encodes a large cell wall-lytic enzyme (71.6 kDa) responsible for staphylococcal peptidoglycan hydrolysis. Phage K is a polyvalent anti-staphylococcal phage with a broad host range. Its lysin LysK gene was cloned and heterologously over-expressed. The recombinant phi 11 endolysin was found to hydrolyze heat-killed staphylococci as well as staphylococcal biofilms [31]. In the present study, we cloned and characterized the LysSA4 gene of S. aureus phage SA4 in order to determine the levels of similarity and heterogeneity with those of other S. aureus phages. Additionally, in vitro lytic activity of the purified recombinant LysSA4 was measured and compared against 68 mastitogenic Staphylococcus isolates of bovine origin to determine the lytic range of the LysSA4 enzyme.
The indicator S. aureus and 68 regional mastitogenic Staphylococcus isolates of bovine origin were either obtained from the Type Culture Laboratory of the Division of Biological Standardization, Indian Veterinary Research Institute (IVRI), India, or were identified during the current study. S. aureus bacteriophage SA4 isolated from the sewage water of an IVRI dairy farm per the recommended method [19] was obtained from the Laboratory Repository of the Division of Biological Standardization, IVRI, India.
The propagation of the bacteriophage was done by conventional liquid culture method. The 100 mL of 16 to 18 h old pure broth culture of the indicator bacterium (S. aureus) with 0.1% MgSO4.7H2O (HiMedia, India) was inoculated with 1.0 mL of the phage suspension. The phage bacteria mixture was incubated at 37℃ with vigorous intermittent shaking until complete lysis (approximately 3 h). The phage lysate was filtered through 0.22 µ membrane filter (EMD Millipore, USA), and stored aseptically in sterile screw capped bottle at 4℃. Genomic DNA of S. aureus bacteriophage SA4 was isolated using a lambda DNA isolation kit (Qiagen, Germany). The LysSA4 gene from the phage was amplified by PCR using specific primers (a kind gift from Dr. D.M. Donovan, United States Department of Agriculture, USA): 5' ATGGCTAAGACT CAAGCAGAA 3' (sense primer) and 5' ATTTACCCGT GTGCCAAG 3' (antisense primer). The PCR reaction consisting of 50 ng of the template DNA, 20 pmol of each primer, and 17 µL of a PCR master mix (Qiagen, Germany) was brought up to a 25-µL volume with nuclease-free water. PCR was performed with an initial denaturation at 94℃ for 5 min followed by 30 cycles of denaturation at 94℃ for 60 sec, annealing at 51.8℃ for 60 sec, extension at 72℃ for 60 sec, and final extension at 72℃ for 7 min. The PCR products were separated in a 1% agarose gel with a 100-bp marker (Thermo Scientific, USA) to determine the gene size.
The LysSA4 gene PCR product was ligated into a pTZ57R/T-TA cloning vector (Qiagen, Germany) and used to transform Escherichia (E.) coli DH5α (Division of Biochemistry, IVRI, India) using a transformation kit (Thermo Scientific, USA). Recombinant clones were preliminarily identified by blue/white colony screening on Luria Bertani agar (HiMedia, India) after overnight incubation at 37℃. Plasmid isolation was carried out using a Plasmid Miniprep kit (Qiagen, Germany) according to the manufacturer's protocol and clone identity was confirmed by digestion at 37℃ overnight with EcoRI (Thermo Scientific, USA). The confirmed clone was then used for sequencing the inserted gene. The gene was sequenced by Chromous Biotech (India) and the sequence was compared with other published lysin sequences in GenBank (National Center for Biotechnology Information, USA) using the ClustalW subprogram of the MegAlign program of Lasergene software (DNASTAR, USA). A multiple sequence alignment report, sequence similarity/divergence data, and phylogenetic tree were generated. Secondary structure predictions and physiochemical properties of LysSA4 in the form of an algorithm were generated with the Protean program (DNASTAR, USA).
The LysSA4 gene was cloned into a pQE30-UA expression vector (Qiagen, Germany) and the endolysin protein was expressed in E. coli M15 competent cells (Division of Biochemistry, IVRI, India) using standard procedures [30]. Briefly, 100 µL of an overnight culture containing the recombinant clones were transferred into 10 mL of Luria Bertani broth (HiMedia, India) containing ampicillin (100 µg/mL; HiMedia, India) and kanamycin (50 µg/mL; HiMedia, India), and incubated in shaker (10 × g; New Brunswick, USA) at 37℃ for 3 h. A 1-mL aliquot of the culture from this stage was taken and kept separately as untreated culture. The remaining cultures were treated with isopropyl β-D-1-thiogalactopyranoside (IPTG; Thermo Scientific, USA) at a final concentration of 1 mM and incubated at 33℃ up to 8 h. A 2-mL aliquot of the culture was harvested every 2 h and the cells were pelleted at 14,000 × g/4℃ for 10 min in a refrigerated microcentrifuge (Sorvall RC5; Thermo Scientific, USA) for SDS-PAGE analysis. The pellet was suspended in 80 µL of 2× sample loading buffer (Merck, USA), boiled for 7 min at 100℃, and centrifuged at 7,000 × g for 10 min at 4℃. The supernatant (20 µL) was loaded with an medium range molecular weight marker [(Merck, USA) and the proteins were separated by minigel SDS-PAGE (Bio-Rad, USA) with a 12% separating gel (Merck, USA) and 5% stacking gel (Merck, USA)]. The time at which the highest level of LysSA4 expression was observed was noted and used as the standard incubation period to achieve a maximum level of expression.
A 50-mL aliquot of the IPTG-treated culture was centrifuged at 4,000 × g for 10 min at 4℃. The cell pellet was resuspended in 2 mL of lysis buffer (100 mM NaH2PO4, 10 mM Tris, and 8 M urea, pH 8.0), and kept at room temperature (RT) for 2 h at 5 × g in a shaker incubator (New Brunswick, USA). The bacterial lysate was centrifuged at 10,000 × g for 20 min at 4℃ to remove the insoluble cellular debris. Supernatant containing the recombinant protein was loaded onto 1 mL of a 50% nickel nitrilotriacetic acid agarose slurry (Qiagen, Germany) and mixed gently by shaking 5 × g for 1 h at RT. Next, the mixture was loaded into a syringe column (HMD, India) and the column was washed with 10 mL of wash buffer (100 mM NaH2PO4, 10 mM Tris, and 8 M urea, pH 6.3) to remove the non-specific proteins. Protein bound to the resin was then eluted with elution buffer (100 mM NaH2PO4, 10 mM Tris, and 8 M urea, pH 4.5) into four eluates of 0.6 µL each. The purified protein was dialyzed against PBS at 4℃ for 48 h. All four eluates were analyzed by SDS-PAGE along with a medium range protein (Merck, USA) to confirm the presence of the protein.
Lytic activity of the protein against its indicator bacteria and the 68 regional mastitogenic Staphylococcus isolates was measured with a spot inoculation test as previously described [25] using the elutes containing protein over lawn cultures of the bacteria. Briefly, each culture of Staphylococcus was inoculated into sterilized BHI broth (HiMedia, India), and incubated at 37℃. The 16 h old pure broth culture of each isolate was spread plated onto the BHI agar (HiMedia, India) and thereafter, 5 µL of the protein preparation was aseptically placed on the dried surface of agar. After overnight incubation at 37℃, the sensitivity of target organism against the phage was observed by formation of clear circular zone.
The lysin gene from phage SA4 was successfully amplified using LysSA4-specific primers. A single 802-bp PCR product was amplified from the phage SA4 genome (Fig. 1A). The amplified product was cloned into a pTZ57R/T-TA cloning vector (Qiagen, Germany). Restriction digestion of the recombinant clone with EcoRI confirmed the presence of the 802-bp sequence within the recombinant plasmid (Fig. 1B).
The cloned LysSA4 gene of S. aureus bacteriophage SA4 was sequenced. The sequence data was submitted to the GenBank database (National Center for Biotechnology Information, USA) and assigned the accession number HQ330981. Sequence analysis of the recombinant clone revealed the presence of a single 802-bp open reading frame. The LysSA4 sequence was then aligned with reported sequences of endolysin genes from other phages. Analysis showed that LysSA4 had a 94.6~97.0% identity with endolysin genes from A5W, G1, GH15, ISP, K, Phi812, and Sb-1 phages (Fig. 2A). Phylogenetic analysis of the nucleotide sequences demonstrated that the endolysin gene of phage SA4 grouped with that of phage GH15 (Fig. 2B), and both of these sequences formed a cluster that was separate from the other sequences.
The deduced amino acid sequence of the LysSA4 gene encoded a partial protein with a calculated molecular mass of 30 kDa. Amino acid sequence of this endolysin gene was then aligned with those of other reported phages. Amino acid sequence analysis indicated that LysSA4 had a 98.5~98.9% similarity with the sequences of endolysin genes from other phages (Fig. 3A). Phylogenetic analysis of the endolysin gene amino acid sequences confirmed the nucleotide sequence analysis findings although the phage SA4 formed a lineage separate from all other reported phages (Fig. 3B). These results indicated that the endolysin genes are conserved among different phages with minor differences.
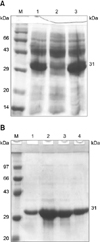
The 802-bp lysin gene product was amplified from the phage SA4 genome and cloned into a pQE30-UA expression vector. Restriction digestion with EcoRI confirmed the presence of an 802-bp sequence within the recombinant plasmid (Fig. 1B). Transformed E. coli M15 cells were then treated with IPTG and the expressed recombinant lysin (Fig. 4A) fused to a histidine tag was purified by nickel chelating affinity chromatography. SDS-PAGE electrophoresis of the eluted fractions showed an intense protein band of 31 kDa (Fig. 4B).
In the spot inoculation test, the expressed endolysin completely cleared the indicator bacterium (S. aureus isolate IVRI-139; Fig. 5). Lytic efficacy against approximately 18% of total Staphylococcus isolates tested in the study was observed (Table 1). The recombinant endolysin also showed cross-species lytic efficacy (Table 1).
The emergence of drug-resistant S. aureus strains necessitates the development of novel therapeutic tools as alternatives for antibiotics. Phage therapy is one such modality that could replace antibiotics. The lysin genes of S. aureus phages have been cloned, sequenced, and characterized by several groups [13,14,24,27]. Recombinant lysins have also been found to be lethal against Lactobacillus lactis [32], Listeria monocytogenes [16], Bacillus cereus [15], S. aureus [14], E. coli [21], and Pseudomonas aeruginosa [21]. In the present study, we cloned the LysSA4 gene and characterized in vitro lytic activity of the recombinant LysSA4 protein against different mastitogenic S. aureus isolates of bovine origin. Sequence analysis of the LysSA4 gene of phage SA4 revealed a single open reading frame encoding a partial protein with a calculated mass of 30 kDa. This analysis also showed that LysSA4 had about a 94~97% identity with endolysin genes from A5W, G1, GH15, ISP, K, Phi812, and Sb-1 phages. Phylogenetic analysis of the nucleotide sequences showed that the endolysin gene of phage SA4 grouped together with that of phage GH15 and formed a separate cluster. However, amino acid sequence analysis indicated that phage SA4 formed a lineage separate from all other reported phages. Our data indicated that the endolysin genes are conserved among different phages with minor differences.
LysSA4 was expressed in E. coli M15 cells and SDS-PAGE showed that the expressed LysSA4 had an apparent molecular mass of 31 kDa. The lytic nature of the recombinant LysSA4 protein was demonstrated by spot inoculation over a lawn culture of indicator organisms. The expressed endolysin was found to have lytic activity against about 18% of the total S. aureus isolates tested. The recombinant endolysin also showed cross-species lytic efficacy. This type of activity demonstrated the therapeutic potential of the expressed lysin. The lytic range of the expressed lysin was found to be low compared to the native lysin [20]. Our results are in agreements with earlier observations [9,11] showing that the native protein provides better protection than recombinant ones.
In the current study, the recombinant LysSA4 protein was found to have only a narrow range for lysing S. aureus isolates compared to the native protein. However, findings from the present study revealed that recombinant LysSA4 could potentially be used as a therapeutic tool. Further research to identify the residues crucial for endolytic activities may help devise peptide-based therapy against S. aureus-induced mastitis.
Figures and Tables
Fig. 1
Cloning of the LysSA4 gene from phage SA4. (A) Amplification of the LysSA4 gene. Lane M: 100-bp plus DNA ladder and lane 1: the amplified product. (B) EcoRI digestion of recombinant vectors harboring the LysSA4 gene. Lane M1: high range DNA ladder, lane M2: 100-bp DNA ladder, lane 1: released gene insert from the recombinant pTZ57R/T-TA cloning vector, and lane 2: released gene insert from the recombinant pQE30-UA expression vector.
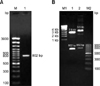
Fig. 2
Nucleotide sequence analyses of the LysSA4 gene from Staphylococcus (S.) aureus phage SA4. (A) Percent identity and divergence of the lysin gene sequences for different phages. (B) Phylogenetic analysis of the lysin sequences for different phages.

Fig. 3
Amino acid sequence analyses of the LysSA4 gene from S. aureus phage SA4. (A) Percent identity and divergence of the lysin protein sequences for different phages. (B) Phylogenetic analysis of the lysin protein sequences for different phages.

Fig. 4
Expression of the recombinant LysSA4 gene from phage SA4. (A) SDS-PAGE showing expression of the LysSA4 gene 4 and 6 h after isopropyl β-D-1-thiogalactopyranoside (IPTG) treatment. Lane M: protein molecular weight marker, lanes 1 and 3: expression 4 and 6 h post-induction, and lane 2: untreated cell lysate. (B) Elution pattern of the purified recombinant lysin from phage SA4. Lane M: protein molecular weight marker (medium range), lanes 1 to 4: eluates 1 to 4. Eluate 2 had the maximum concentration of the protein followed by eluates 3, 4, and 1, respectively.
References
1. Andersson I, van Scheltinga ACT, Valegård K. Towards new β-lactam antibiotics. Cell Mol Life Sci. 2001. 58:1897–1906.

2. Bishop EJ, Howden BP. Treatment of Staphylococcus aureus infections: new issues, emerging therapies and future directions. Expert Opin Emerg Drugs. 2007. 12:1–22.

3. Bon J, Mani N, Jayaswal RK. Molecular analysis of lytic genes of bacteriophage 80α of Staphylococcus aureus. Can J Microbiol. 1997. 43:612–616.

4. Borysowski J, Weber-Dąbrowska B, Górski A. Bacteriophage endolysins as a novel class of antibacterial agents. Exp Biol Med (Maywood). 2006. 231:366–377.

5. DeGraves FJ, Fetrow J. Economics of mastitis and mastitis control. Vet Clin North Am Food Anim Pract. 1993. 9:421–434.

6. Deluyker HA, Chester ST, Van Oye SN. A multilocation clinical trial in lactating dairy cows affected with clinical mastitis to compare the efficacy of treatment with intramammary infusions of a lincomycin/neomycin combination with an ampicillin/cloxacillin combination. J Vet Pharmacol Ther. 1999. 22:274–282.

7. Donovan DM. Bacteriophage and peptidoglycan degrading enzymes with antimicrobial applications. Recent Pat Biotechnol. 2007. 1:113–122.

8. Doughty CC, Mann JA. Purification and properties of a bacteriophage-induced cell wall peptidase from Staphylococcus aureus. J Bacteriol. 1967. 93:1089–1095.

9. Etlinger HM, Caspers P, Matile H, Schoenfeld HJ, Stueber D, Takacs B. Ability of recombinant or native proteins to protect monkeys against heterologous challenge with Plasmodium falciparum. Infect Immun. 1991. 59:3498–3503.

10. Gill JJ, Pacan JC, Carson ME, Leslie KE, Griffiths MW, Sabour PM. Efficacy and pharmacokinetics of bacteriophage therapy in treatment of subclinical Staphylococcus aureus mastitis in lactating dairy cattle. Antimicrob Agents Chemother. 2006. 50:2912–2918.

11. Heath ACG, Broadwell AH, Chilcott CN, Wigley PJ, Shoemaker CB. Efficacy of native and recombinant Cry1B protein against experimentally induced and naturally acquired ovine myiasis (fly strike) in sheep. J Econ Entomol. 2004. 97:1797–1804.

12. Loeffler JM, Fischetti VA. Synergistic lethal effect of a combination of phage lytic enzymes with different activities on penicillin-sensitive and -resistant Streptococcus pneumoniae strains. Antimicrob Agents Chemother. 2003. 47:375–377.

13. Loessner MJ, Gaeng S, Scherer S. Evidence for a holin-like protein gene fully embedded out of frame in the endolysin gene of Staphylococcus aureus bacteriophage 187. J Bacteriol. 1999. 181:4452–4460.

14. Loessner MJ, Gaeng S, Wendlinger G, Maier SK, Scherer S. The two-component lysis system of Staphylococcus aureus bacteriophage Twort: a large TTG-start holin and an associated amidase endolysin. FEMS Microbiol Lett. 1998. 162:265–274.

15. Loessner MJ, Maier SK, Daubek-Puza H, Wendlinger G, Scherer S. Three Bacillus cereus bacteriophage endolysins are unrelated but reveal high homology to cell wall hydrolases from different bacilli. J Bacteriol. 1997. 179:2845–2851.

16. Loessner MJ, Wendlinger G, Scherer S. Heterogeneous endolysins in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. Mol Microbiol. 1995. 16:1231–1241.

17. Lorch A. Bacteriophages: An alternative to antibiotics? Biotechnol Dev Monit. 1999. 39:14–17.
19. Markel DE, Eklund C. Isolation, characterization, and classification of three bacteriophage isolates for the genus Levinea. Int J Syst Bacteriol. 1974. 24:230–234.

20. Mishra AK, Rawat M, Abhishek , Sureshkannan . Characterization and lytic activity of endolysin induced by bacteriophage SA4 against mastitogenic isolates of Staphylococcus of bovine origin. Indian Vet J. 2012. 89:24–26.
21. Morita M, Tanji Y, Orito Y, Mizoguchi K, Soejima A, Unno H. Functional analysis of antibacterial activity of Bacillus amyloliquefaciens phage endolysin against Gram-negative bacteria. FEBS Lett. 2001. 500:56–59.

22. Navarre WW, Ton-That H, Faull KF, Schneewind O. Multiple enzymatic activities of the murein hydrolase from staphylococcal phage φ11. Identification of a D-alanyl-glycine endopeptidase activity. J Biol Chem. 1999. 274:15847–15856.

23. Nelson D, Loomis L, Fischetti VA. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc Natl Acad Sci USA. 2001. 98:4107–4112.

24. O'Flaherty S, Coffey A, Meaney W, Fitzgerald GF, Ross RP. The recombinant phage lysin LysK has a broad spectrum of lytic activity against clinically relevant staphylococci, including methicillin-resistant Staphylococcus aureus. J Bacteriol. 2005. 187:7161–7164.
25. Park SC, Shimamura I, Fukunaga M, Mori KI, Nakai T. Isolation of bacteriophages specific to a fish pathogen, Pseudomonas plecoglossicida, as a candidate for disease control. Appl Environ Microbiol. 2000. 66:1416–1422.

26. Paul VD, Rajagopalan SS, Sundarrajan S, George SE, Asrani JY, Pillai R, Chikkamadaiah R, Durgaiah M, Sriram B, Padmanabhan S. A novel bacteriophage tail-associated muralytic enzyme (TAME) from phage K and its development into a potent a antistaphylococcal protein. BMC Microbiol. 2011. 11:226.
27. Ralston DJ, Baer BS, Lieberman M, Krueger AP. Virolysin: a virus-induced lysine from staphylococcal phage lysates. Proc Soc Exp Biol Med. 1955. 89:502–507.
28. Rodríguez L, Martínez B, Zhou Y, Rodríguez A, Donovan DM, García P. Lytic activity of the virion-associated peptidoglycan hydrolase HydH5 of Staphylococcus aureus bacteriophage vB_SauS-phiIPLA88. BMC Microbiol. 2011. 11:138.
29. Rodríguez-Rubio L, Gutiérrez D, Martínez B, Rodríguez A, García P. Lytic activity of LysH5 endolysin secreted by Lactococcus lactis using the secretion signal sequence of bacteriocin Lcn972. Appl Environ Microbiol. 2012. 78:3469–3472.

30. Sambrook J, Russell DW. Molecular Cloning: a Laboratory Manual. 2001. Vol. 3:3rd ed. New York: Cold Spring Harbor Laboratory Press;15.14–15.18.
31. Sass P, Bierbaum G. Lytic activity of recombinant bacteriophage φ11 and φ12 endolysins on whole cells and biofilms of Staphylococcus aureus. Appl Environ Microbiol. 2007. 73:347–352.

32. Shearman C, Underwood H, Jury K, Gasson M. Cloning and DNA sequence analysis of a Lactococcus bacteriophage lysin gene. Mol Gen Genet. 1989. 218:214–221.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download