Abstract
Molecular mechanisms governing peritonitis caused by the presence of aseptic gauze have remained unclear. To identify the genes involved, sterile gauze-exposed omentum was collected at 0, 6, 12, 24, and 48 h intervals, and analyzed by differential display RT(reverse transcription)-PCR. Among over 1,200 bands, 230 bands were found differentially expressed. These bands represented the fragment sizes of approximately 200 to 1,500 bp. The eight fragments were expressed differentially in the treatment group but not in the control. The sequences of two bands were similar to those of genes associated with the inflammatory process and a band was related to repair and regeneration process. Another one was related with spermatogonia and the rest four were unknown. Additionally, amplicons corresponding to the full-length sequences of two inflammatory gene fragments were synthesized by rapid amplification of cDNA end PCR. One showed 99% similarity to the major histocompatibility complex class II dog leukocyte antigen-DR beta chain and the other was canis familiaris proteasome beta type 3. Results of the present study suggested that sterile gauze induced the differential expression of genes in the omentum involved in inflammation and healing process.
Tissue damage initiates a cascade of inflammatory and matrix remodeling events that are necessary to restore tissue integrity and function [1]. Activation of the inflammatory system is commonly influenced by up-regulation of proinflammatory mediators at the mRNA level [17]. The omentum is the source of fibroblasts, and contains lymphatic vessels, lymph nodes, and nerves. This unique tissue responds to inflammation [18], adheres firmly to the site of wound closure [7], and participates in healing. Gauze is the most frequently retained foreign body in the abdomen [16] because of its common usage, small size, and amorphous structure. The retained gauze evokes an inflammatory reaction and becomes surrounded by omentum.
Differential display reverse transcription polymerase chain reaction (DD RT-PCR) is a powerful technique for identifying different sets of genes expressed in specific tissues or cell types [9]. It does not require any prior mRNA genomic information and is possible to identify novel and unexpected changes in transcription, making the non-biased gene screening feasible [11].
Pathophysiological and molecular mechanisms underlying peritonitis caused by microbiological and chemical agents have been elucidated [15], but the molecular mechanism governing peritonitis caused by aseptic gauze has remained elusive. The aim of the present study was to examine the early inflammatory process initiated by gauze introduced into the omentum and identify changes in gene expression.
Three healthy beagle dogs ranging from 2 to 3 years old with a mean weight of 4.5 kg were used for this study. All procedures were performed according to the "Guide for the Care and Use of Laboratory Animals"of Seoul National University (Korea). Anesthesia was induced with propofol (Provive 1%; Claris Lifesciences, India) and was maintained with 2% isoflurane (Terrell; Minrad International, USA). An aseptic incision was made in abdomen and pieces of sterile gauze were packed into the abdomen of two dogs in the treated group and a control dog underwent a sham operation. Omental samples were taken from the dog at 0, 6, 12, 24, and 48 h after the operation. After 48 h, the gauze was removed from the abdomen, the abdominal cavity was flushed with saline solution, and the wound was closed with sutures. Omental tissues were cut into multiple pieces less than 0.5 cm thick and immediately submerged in an RNA stabilization reagent (RNAlater; Qiagen, Germany).
Total RNA from the omentum specimens was isolated using a modified acid-guanidine thiocyanate-phenolchloroform method with TRIzol reagent (Invitrogen, USA) according to the manufacturer's instructions. RNA quality was verified by staining rRNA bands with ethidium bromide in a 1.5% agarose mini gel, and ensuring that the OD 260/280 ratios were between 1.8 and 2.0 using spectrophotometer (Smartspec 3000; Bio-Rad, USA). RT was conducted by the reagents of GeneFishing DEG Premix Kit (Seegene, Korea) except reverse transcriptase SuperScript II (Invitrogen, USA). Three µg of total RNA was mixed with 2 µL of 10 mM cDNA synthesis primer dT-ACP1 (Seegene, Korea) and DEPC-treated water to make the total volume of 9.5 µL. The mixture was incubated at 80℃ for 3 min, and immediately placed on ice for 2 min before being briefly centrifuged. After that, the mixture was added by 4 µL of 5× RT buffer (Invitrogen, USA), 4 µL of 2.5 mM dNTPs, 20 units of 0.5 µL RNase inhibitor, and 200 units of 1 µL reverse transcriptase with a final volume of 20 µL. RT was performed at 42℃ for 90 min and the reaction was inactivated at 94℃ for 2 min. After being incubated on ice for 2 min, the reaction was diluted five-fold by adding 80 µL of distilled water and stored at -20℃ before further use.
Second-strand cDNA synthesis and further amplification were conducted in a single tube as per the manufacturer's instructions (GeneFishing DEG Premix Kit; Seegene, Korea). Second-strand cDNA synthesis was conducted at 50℃ for one cycle of first-stage PCR in a final reaction volume of 20 µL containing 3 µL of the diluted first-strand cDNA, 10 µL of 2× SeeAmp ACP master mix, 1 µL of 10 mM dT-ACP2 (anchor oligo-dT primer), and 2 µL of 5 µM arbitrary primers of 60 APS and primers for the 8 fragments are presented in Table 1. PCR for second-strand synthesis was performed in one cycle of incubation at 94℃ for 5 min, 50℃ for 3 min, and 72℃ for 1 min. After the completion of second-strand cDNA synthesis, 40 cycles of amplification were performed with denaturation at 94℃ for 40 sec, annealing at 65℃ for 40 sec, and extension at 72℃ for 40 sec; this was followed by a final extension step of 5 min at 72℃.
PCR products were separated in 2% agarose gel with 0.5× TBE buffer at 100 V in mini gel migration trough. Differential gene expression was assessed with PCR samples corresponding to each time interval (0, 6, 12, 24, and 48 h after gauze placement) for both the control and gauze-treated groups. Differences in gene expression were detected by noting whether the intensities of bands corresponding to the same RNA fragment varied across the different lanes. Bands that did not change in intensity for the control group but altered in the treated group were considered to represent candidate genes associated with responses to gauze introduction. Differentially expressed cDNA bands were excised from the gels and purified.
Differentially expressed bands were processed with a MEGA-spin agarose gel extraction kit (iNtRON Biotechnology, Korea). Briefly, the bands of interest were excised using a sharp blade from the gel on a UV-illuminator. Agarose lysis buffer of 60 µL was added for 10 mg of gel. The gel pieces were incubated at 50℃ for 10 min and mixed by vortexing during this time. The melted gel was transferred to a DNA-binding column with a 2 µL collection tube, and centrifuged for 1 min at 15,000 G. The column was washed with washing buffer and centrifuged, and the bound DNA from the band was recovered with elution buffer.
The eluted cDNA bands were ligated into pGEM-T plasmid (Promega, USA) and used to transform into DH5α (Escherichia coli) competent cells (Invitrogen, USA) using standard cloning protocols. The clones were sequenced bi-directionally using T7 forward and SP6 reverse primers (National Instrumentation Center for Environmental Management, Korea). All sequences were analyzed using BLAST and FASTA with GenBank (National Center for Biotechnology Information, USA) and EMBL (Germany) databases to determine sequence identity and tissue distribution. The alignment of forward and backward nucleotide sequences was performed by aligning two sequences (bl2seq), and nucleotide sequence homology was evaluated by nucleotide-nucleotide BLAST (blastn).
RACE was performed with commercially available kits (CapFishing Full-length cDNA Premix Kit, Seegene, Korea) to obtain 5'side-overlapping cDNA from the candidate clones. To generate full-length cDNA transcripts, 3 µg of total RNA were reverse-transcribed using a universal oligo(dT)-adaptor primer. PCR amplification was performed with a combination of the universal primer and target-specific primers (TSP). The primer combinations used were: 5'-RACE Primer (forward): 5'GTC TAC CAG GCA TTC GCT TCA T 3', TSP (reverse): 5' GGG CTG CAT AGG AAG AGG AAG GAG AG 3' for clone 250-48, and 5'-RACE Primer (forward): 5' GTC TAC CAG GCA TTC GCT TCA T 3', TSP (reverse): 5'GCC TCG TCC ACG GTG ACT GGT GTT TC 3' for clone 400-46. RACE amplification using the modified adaptor primers was performed to obtain both ends of the full-length gene. PCR was performed at 94℃ for 3 min for the initial denaturation followed by 30 cycles of 94℃ for 40 sec, 66℃ for 40 sec, and 72℃ for 1 min with a final extension at 72℃ for 5 min for clone 250-48. The same PCR was used for the full-length synthesis of clone 400-46 except annealing temperature (63℃) differed. The PCR products were separated in 2% agarose gels and the band size was preliminarily confirmed by comparison with a DNA ladder. The obtained DNA sequences were compared to genes in the GenBank database.
A total of 60 PCRs for each time of each animal were performed. Approximately 6~10 bands were detected with each pair of PCR primers in each animal. Thus, over 1,200 RNA species were analyzed. Among them, 230 bands were found differentially expressed compared to their respective time course intervals. These bands represented the fragment sizes of 200 to 1,500 bp. A representative band indicating differential expression between the control and gauze-implanted groups is shown in Fig. 1. Eight bands were expressed differentially only in the treatment group but not in the control. Nine bands were altered in both groups but more high or less intensity in the treatment group.
Presumptive functions and characteristics of genes corresponding to eight fragments expressed differentially only in the treatment group were determined based on sequence similarities with genes found in the databases we used (Table 2). Clone 250-48 showed 99% sequence homology to major histocompatibility complex (MHC) class II dog leukocyte antigen (DLA)-DR-beta chain. Homologies (99~100%) were also discovered between 400-29 and canis familiaris DAZ interacting protein 1, mRNA; clone 400-46 and canis familiaris proteasome beta type 3; and clone 750-47 and predicted heterogenous nuclear ribonuclear protein (hnRNP) H according to our GenBank-based analysis. Clones 300-22, 400-20, 600-14 and 900-32 did not have significant sequence homology with those of existing genes in GenBank. Clones 250-48 and 400-46 were found to have sequence similarities with genes related to inflammatory processes whereas clone 750-47 appears to correspond to gene that influence development and repair processes in the body. Gene accession numbers and lengths of the sequences are shown in Table 3.
Full-length sequence of inflammatory gene fragments 250-48 and 400-46 were synthesized by 5'-RACE PCR (Fig. 2). The full-length cDNA sequence of clone 250-48 was 1243 bp long and that of clone 400-46 was 781 bp (Fig. 3). The sequence of clone 250-48 contained an ATG start codon and TGA stop codon with an open reading frame (ORF) of 802 bp. The up-stream length of the gene was 96 bp and the down-stream length was 315 bp. A consensus sequence (AATAAA) was found 16 bp up-stream of the poly(A) tail. This full-length sequence was 99% homologous to that of the canine MHC class II DLA-DR-beta chain gene. The sequence of clone 400-46 contained an ATG and TAA with an ORF of 619 bp and a consensus sequence (AATAAA) was found 13 bp up-stream of the poly(A) tail. The full-length sequence of clone 400-46 showed a 99% homology with that of the canis familiaris proteasome beta type 3 gene.
An annealing control primer-based PCR system used in this study is a modification of the original DD RT-PCR [12] by changing the primer design strategy. Annealing control oligo-dT primers (dT-ACP1 and dT-ACP2) were used for the synthesis of first-strand cDNA and subsequent PCR amplification to ensure accurate annealing [6]. DD RT-PCR is a laborious method with high rate of false positives and redundancy [21] and the downstream screening steps constitute major limitations [14]. However, the novel oligonucleotide primer designated 'Annealing Control Primer' (ACP) provides a high annealing specificity to the target sequences and allows only genuine products to be amplified [6]. Identifications of differentially expressed genes in other diseases using annealing control primer (ACP)-based GeneFishing PCR were reported [10,13]. Thus, the gene fragments generated from samples of omentum with gauze implantation are supposed to be accurately produced by our ACP-based DD RT-PCR technique.
MHC class II DLA-DR beta chain is involved in the initiation of inflammation because it induced severe inflammation in corneal endothelium and joints [8,20]. MHC class II molecules are constitutively expressed on cells that serve as antigen presenting cells for CD4+ T cells, such as macrophages, monocytes, dentritic cells [19]. The proteasome mediates the degradation of most short-lived proteins that control cell cycle, transcription, DNA repair, apoptosis, and other cellular processes [4].
Expressional changes in pro-and anti-inflammatory cytokines and antibacterial peptides in the normal human omentum when exposed lipopolysaccharide were demonstrated as compared with oral mucosa [3]. Although expressions of differential genes could not be compared between omentum and other tissues in the present study, differential genes were derived from the omentum which is severely inflamed by gauze implanted. These genes could be used to early markers for peritonitis. Heterogeneous nuclear ribonucleoprotein (hnRNP) H related to repair and regeneration processes. hnRNP plays a pivotal role in coordinating repair pathways following exposure to ionizing radiation [5]. Canis familiaris DAZ interacting protein 1 is present predominantly in spermatogonia related with germ cell maturation [2]. It was not considered that DAZ is related with omentum or peritonitis.
In the present study we cloned two inflammatory related genes from gauze implanted omentum, canis familiaris MHC class II DLA-DR beta chain and proteasome beta type 3 by DD RT-PCR and RACE PCR. Further studies are needed to evaluate unknown genes and for the use as early markers for peritonitis with known genes.
Figures and Tables
Fig. 1
Representative images of the annealing control primer-based PCR products for identifying differentially expressed genes during inflammation induction in cases of canine peritonitis. RNA fingerprinting results for the control and treatment animals at 0, 6, 12, 24, and 48 h with GAPDH as the internal control. Amplified cDNA products correspond to gene transcripts in the omentum associated with the response to gauze implantation. Lane M: 100 bp size marker. The right arrow indicates bands that correspond to changes in cDNA expression. The cDNA band was excised from the gel for further cloning and sequencing.
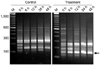
Fig. 2
Bands generated by 5'end rapid amplification of cDNA end (RACE) for clones 250-48 and 400-46. (A) Clone 250-48. (B) Clone 400-46. PCR products were separated in a 2% agarose gel. Lane 2 in each gel contains the 5'RACE PCR products; Lane M: DNA ladder. The arrows indicate the 5'RACE products in the gels.
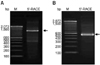
Fig. 3
The sequence of the 5'end for clones 250-48 and 400-46 synthesized from the omentum of dogs along with the full-length sequences are shown. The full-length sequence of clone 250-48 synthesized from the canine omentum is shown in (A) and that of clone 400-46 appears in (B). The underlined sequences correspond to annealing sites of the forward primers used for ACP-based full-length gene synthesis (5'RACE primer). The ATG start codon and TAA stop codon are shaded, and the boxed consensus sequence AATAA is the signal for polyadenylation.

Table 1
Primer sequences used for cDNA synthesis and annealing control primer (ACP)-based PCR for examining altered gene expression in canine omentum with peritonitis

Acknowledgments
The authors are grateful for the financial support from the Research Institute for Veterinary Science, College of Veterinary Medicine, Seoul National University, Korea.
References
1. Azouz A, Razzaque MS, El-Hallak M, Taguchi T. Immunoinflammatory responses and fibrogenesis. Med Electron Microsc. 2004; 37:141–148.

2. Bush JM, Gardiner DW, Palmer JS, Rajpert-De Meyts E, Veeramachaneni DNR. Testicular germ cell tumors in dogs are predominantly of spermatocytic seminoma type and are frequently associated with somatic cell tumors. Int J Androl. 2011; 34(4 Pt 2):e288–e295.
3. Chandra A, Srivastava RK, Kashyap MP, Kumar R, Srivastava RN, Pant AB. The anti-inflammatory and antibacterial basis of human omental defense: selective expression of cytokines and antimicrobial peptides. PLoS One. 2011; 6:e20446.

4. Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002; 82:373–428.

5. Haley B, Paunesku T, Protic M, Woloschak GE. Response of heterogenous ribonuclear proteins (hnRNP) to ionising radiation and their involvement in DNA damage repair. Int J Radiat Biol. 2009; 85:643–655.

6. Hwang IT, Kim YJ, Kim SH, Kwak CI, Gu YY, Chun JY. Annealing control primer system for improving specificity of PCR amplification. Biotechniques. 2003; 35:1180–1184.

7. Jani K, Saxena AK, Vaghasia R. Omental plugging for large-sized duodenal peptic perforations: a prospective randomized study of 100 patients. South Med J. 2006; 99:467–471.

8. Kanazawa S, Ota S, Sekine C, Tada T, Otsuka T, Okamoto T, Sønderstrup G, Peterlin BM. Aberrant MHC class II expression in mouse joints leads to arthritis with extraarticular manifestations similar to rheumatoid arthritis. Proc Natl Acad Sci U S A. 2006; 103:14465–14470.

9. Kao RH, Francia G, Poulsom R, Hanby AM, Hart IR. Application of differential display, with in situ hybridization verification, to microscopic samples of breast cancer tissue. Int J Exp Pathol. 2003; 84:207–212.

10. Kim YS, Do JH, Bae S, Bae DH, Ahn WS. Identification of differentially expressed genes using an annealing control primer system in stage III serous ovarian carcinoma. BMC Cancer. 2010; 10:576.

11. Liang P, Averboukh L, Pardee AB. Distribution and cloning of eukaryotic mRNAs by means of differential display: refinements and optimization. Nucleic Acids Res. 1993; 21:3269–3275.

13. Maimunah M, Froemming GA, Nawawi H, Nafeeza MI, Effat O, Rosmadi MY, Mohamed Saifulaman MS. Identification of differentially expressed gene (DEG) in atherosclerotic lesion by annealing control primer (ACP)-based Genefishing™PCR. World Acad Sci Eng Technol. 2012; 67:194–199.
14. Nagasaka T, Boulday G, Fraser CC, Coupel S, Coulon F, Tesson L, Heslan JM, Soulillou JP, Charreau B. Rapid selection of differentially expressed genes in TNFα-activated endothelial cells. Mol Med. 2002; 8:559–567.

16. Risher WH, McKinnon WM. Foreign body in the gastrointestinal tract: intraluminal migration of laparotomy sponge. South Med J. 1991; 84:1042–1045.
17. Risom L, Møller P, Loft S. Oxidative stress-induced DNA damage by particulate air pollution. Mutat Res. 2005; 592:119–137.

18. Schreiber S, Gehrckens A, Raedler A. Activation of the major omentum-associated lymphoid tissue in Crohn disease. Zentralbl Chir. 1995; 120:135–144.
19. Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002; 109:Suppl. S21–S33.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download