Abstract
Despite the development of new technologies, new challenges still remain for large scale proteomic profiling when dealing with complex biological mixtures. Fractionation prior to liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis is usually the preferred method to reduce the complexity of any biological sample. In this study, a gel LC-MS/MS approach was used to explore the stage specific proteome of Cryptosporidium (C.) parvum. To accomplish this, the sporozoite protein of C. parvum was first fractionated using SDS-PAGE with subsequent LC-MS/MS analysis. A total of 135 protein hits were recorded from 20 gel slices (from same gel lane), with many hits occurring in more than one band. Excluding all non-Cryptosporidium entries and proteins with multiple hits, 33 separate C. parvum entries were identified during the study. The overall goal of this study was to reduce sample complexity by protein fractionation and increase the possibility of detecting proteins present in lower abundance in a complex protein mixture.
The introduction of powerful and rapid multidimensional separation and characterization methods has enabled more extensive investigation of systems at the molecular level. Proteomics is a leading technology for the high-throughput analysis of proteins on a genome-wide scale. Cryptosporidium (C.) spp. are important zoonotic parasites causing widespread diarrhoeal disease in man and animals [11,16]. With the completion of genome sequencing projects of C. parvum and C. hominis, the global proteome analysis is now more feasible than before [14,15]. Because two-dimensional electrophoresis (2-DE) has a number of limitations, alternative approaches are essential to explore of the insoluble proteome, which includes various hydrophobic and membrane proteins of C. parvum. Therefore, in this study, an attempt was made to separate C. parvum sporozoite proteins using conventional one-dimensional (1D) SDS-PAGE followed by the excision of contiguous gel slices, each of which was then subjected to in-gel tryptic digestion and tandem MS (MS/MS) analysis (LC-MS/MS analysis).
Despite the development of new technologies, new challenges still remain for large scale proteomic profiling when dealing with complex biological mixtures. Fractionation prior to LC-MS/MS analysis is usually preferred to reduce the complexity of any biological sample. While Multidimensional Protein Identification Technology (MudPIT) and other multidimensional separation strategies have been reported for a number of parasitic organisms [4,6,13], no reports are available regarding the use of this approach to explore the proteome of Cryptosporidium. Therefore, this study was conducted to analyze the Cryptosporidium proteome through reducing sample complexity by protein fractionation, thereby increasing the possibility of low-abundant protein identification from a complex protein mixture. In addition to resolving many of the insoluble proteins (and those not amenable by conventional 2-DE analysis), this approach was designed to complement other gel-based proteomic approaches.
Unless otherwise specified, all chemicals and reagents were purchased from Sigma (Poole, Dorset, UK). Ampholytes and 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfo nate were purchased from BioRad Ltd. (Hertfordshire, UK). Modified porcine trypsin was purchased from Promega (Madison, USA). High performance liquid chromatography (HPLC) grade acetonitrile, HPLC grade methanol, and glacial acetic acid were purchased from Fisher Scientific (UK).
Oocysts of C. parvum passaged in lambs (IOWA strain) were purchased from the Moredun Research Institute (MRI, Scotland). The parasite oocyst suspension was stored at 4℃ in the presence of 1,000 U per mL penicillin and 1,000 µg per mL streptomycin. Excystation of oocysts of C. parvum was then performed as previously described [14]. Briefly, excystation was performed at 37℃ using deoxycholate (DOC) and sodium hydrogen carbonate, which was continued until >80% excystation was observed upon microscopic examination at 400× magnification (~ 2 h). Excystation mixtures were pelleted at 13,000 × g for 1 min, washed with 1 mL of PBS and repelleted at 13,000 × g for 3 min at 4℃. Pellets were used immediately or stored at -80℃.
For 1D-SDS-PAGE, frozen sporozoite pellets were disrupted in 40 µL of gel loading buffer containing 50 mM Tris Hydrochloride (pH 6.8), 100 mM DTT, 2% (w/v) SDS, 0.1% (w/v) bromophenol blue and 10% glycerol. The mixture was boiled at 100℃ for 10 min and then chilled on ice before loading into the SDS-PAGE gel lane. A standard broad range protein molecular weight marker (Cat. no. #RPN5800; Amersham, UK) was used as a ladder in a separate lane.
Polyacrylamide gels (12%) were prepared using a mini gel apparatus (BioRad, USA). The resolving gel was prepared using several constituents including 30% acrylamide in 1.5M Tris-HCl (pH 8.8), 10% (w/v) SDS, 10% (w/v) ammonium persulphate and 10 µL TEMED. The stacking gel (4%) was prepared using 30% acrylamide in 1.5M Tris-HCl (pH 6.8), 10% (w/v) SDS, 10% (w/v) ammonium persulphate (APS) and 5 µL TEMED. The SDS electrophoresis buffer was prepared by dissolving 25 mM Tris-base, 192 mM glycine and 0.1% (w/v) SDS in double distilled deionised water. Separation was performed by electrophoresis at 120 V for 2 h. Gels were then stained by the Coomassie Brilliant blue or Colloidal Coomassie staining technique as previously described [14].
The coomassie stained gels were destained by several washes in distilled water until the background became clear. Protein bands were then excised for in gel digestion with trypsin. Usually, a small portion from the middle of a gel band was sufficient, especially for stronger bands. This was likely because small portions required less acrylamide in the digest and improved diffusion of reagents into and peptides out of the gel slice. For this experiment, the entire gel lane was analyzed, regardless of whether the bands were mild or strong. After the gel slices were excised, they were cut into several pieces, placed into a 1.5 mL eppendorf tube and washed for 1 h in 500 µL of 100 mM ammonium bicarbonate. Next, 10 µL of 45 mM DTT in 150 µL of 100 mM ammonium bicarbonate, which was sufficient to cover the gel pieces, was added, and the proteins were reduced for 30 min at 60℃. After cooling to room temperature, the DTT solution was replaced with 10 µL of 100 mM iodoacetamide and incubated for 30 min at ambient temperature in the dark with occasional vortexing. The gel pieces were then washed with 500 µL of 50% acetonitrile/100 mM ammonium bicarbonate while shaking for 1 h. After discarding the wash, 50 µL of acetonitrile was added to shrink the gel pieces. As the gel pieces turned white, the liquid was removed within 10 min and further dried in a vacuum centrifuge by spinning for 10 min. The gel pieces were then reswollen in 0.2 µg trypsin (Sigma-Aldrich, USA) in 10 µL of 25 mM ammonium bicarbonate. After 15 min, 20 µL of 25 mM ammonium bicarbonate was added to cover gel pieces and keep them wet. For enzymatic cleavage, the sample was incubated at 37℃ overnight in a flatbed shaker. After digestion, the peptides were collected for MS analysis.
The peptides generated from each 1D gel band were separately subjected to LC-MS/MS using a capillary HPLC system and a Q-STAR mass spectrometer (Applied Biosystems, USA). The Q-STAR MS analyzer used in this study contained two analyzers separated by a collision cell. PMF data were recorded in the first analyzer and the high intensity peptides were selected for further analyses by fragmentation in the collision cell. Thereafter, the mass of the fragment ions was measured in the second analyzer, which gave rise to peptide sequence information. For tandem MS (MS/MS) analysis, a similar digestion protocol was followed as for MALDI ToF analysis except that TFA solution was replaced by formic acid (10%). One microliter of this sample solution was injected into a 20 µL injection loop and transferred onto a 5 mm C18 trap column (Dionex pepmap; Thermo Scientific, USA). The elute was sprayed directly into the ion source at the same flow rate. The capillary voltage was set to enable spraying of the sample (nano-electro spray) and the spectra were acquired automatically.
To acquire the MS/MS data, a ToF scan was run for 10 sec to detect double, triple and quadruple charged ions that had mass charge ratios (m/z) between 400 and 1,000. Peptide sequencing was performed by data-directed analysis in which the software was set to switch to MS/MS mode as soon as a double, triple or quadruple charged ion was detected. Each full scan mass spectrum generated by the ToF MS was followed by collision-induced dissociation of the eight most intense parent ions with a signal that reached a threshold of 20 counts/s. The data acquisition was performed using the dynamic exclusion of m/z ratios of already fragmented ions while the fragmentation was performed in positive polarity using nitrogen as the collision gas. Mass spectra were acquired for 15 sec in the range of 50 to 2,000 m/z. For protein identification, the data files containing all MS/MS spectra information were interpreted using the Mascot search engine to search against the NCBInr database and a locally downloaded Cryptosporidium database to identify the genes encoding the particular protein.
The local C. parvum database was constructed by downloading all available sequence information from ftp sites of the CryptoDB database version 3.1 [8], which includes all genome sequences of C. parvum and C. hominis isolates generated at the University of Minnesota and Virginia Commonwealth University/Tufts University School of Veterinary Medicine, USA. This database also contains all sequences submitted to GenBank and EST sequences from the C. parvum EST project at UCSF, California, USA.
After chromatographic analysis, the resulting data set was collected from the mass spectrometer and the MASCOT search tool [12] was used to interpret the tandem mass spectra generated in each step by matching the acquired tandem mass spectra with that predicted from a corresponding sequence database. All Mascot searches were performed using the following search parameters: Database: NCBInr, Taxonomy: alveolata, Enzyme: trypsin, Protein mass range: 1 to 100 kDa, Tolerance: 50 ppm, Missed cleavage: 1, Fixed modification: Carbamidomethylation, Variable modification: Oxidation of methionine, Charge state: MH+. In accordance with the cut-off score provided by MASCOT, the protein was considered to be tentatively identified if a significant score was achieved. The searches were carried out against genomic and protein databases including the NCBInr database [17] and Cryptosporidium genome database and EST database [8]. The C. parvum genome and EST sequences were made available by locally downloaded databases.
The MASCOT algorithm was used to interpret the MS/MS spectra derived during this study. In MASCOT, the score for an MS/MS match is based on the absolute probability (p) that the observed match between the experimental data and the database sequence is not a random event. Therefore, the protein score in a Peptide Summary page is derived from the ion scores and provides a logical order to the report.
A previously described sample preparation protocol was followed for the extraction of proteins from 107 excysted oocysts of C. parvum. One dimensional SDS-PAGE was run using the whole sample volume (containing ~60 µg protein) (Fig. 1A). After visualization by colloidal coomassie staining, whole gel lanes were cut into 20 contiguous slices (Fig. 1B) and each slice was subjected to in-gel tryptic digestion using a modified version of the method described before.
All 20 gel bands excised from the coomassie-stained 1D-SDS gel were analyzed by ESI-quadruple tandem mass spectrometry (ESI-MS/MS). The peptide fragmentation data were searched against a non redundant NCBI database with the help of MASCOT [12]. A complete list of all statistically significant protein hits that was revealed from individual gel bands after a MASCOT search against the NCBInr protein database is provided in appendix I. Bands 5, 10, 18 and 20 did not provide any significant protein hits. A total of 135 protein hits were recorded from the remaining 16 bands, with many occurring in more than one band. Excluding all non-Cryptosporidium entries and proteins with multiple hits, 33 separate Cryptosporidium entries were identified (Table 1). Fig. 2 shows the number of redundant hits in each SDS-PAGE gel band. Several matches were to other Apicomplexa and microbial proteins, suggesting a close homology with Cryptosporidium proteins. Band 1 was found to give the largest number of hits (n = 22) hits, with 50% corresponding to Cryptosporidium entries. Several Cryptosporidium proteins including a135 protein (gi.20513131), actin (gi.323089), elongation factor 1 alpha (gi.2894790), oocyst wall protein (gi.46226838), protein disulphide isomerase (gi.32398654) and glyceraldehyde-3-phosphate isomerase (gi.46229140) were reliably identified with high MASCOT scores. The search also indicated the presence of three hypothetical proteins of C. parvum (gi.46229263, gi.32399103, gi.32399022) and one ribosomal protein S5 of C. hominis (gi.54656279). A number of hypothetical proteins of Plasmodium falciparum 3D7 (gi.23613051, gi.23508281, gi.23619138, gi.23619234) were also found, indicating the presence of homologous hypothetical proteins in C. parvum. Other significant hits included subtilisin like protease of Toxoplasma gondii (gi.29378311), myosin D of Plasmodium yoelii (gi.23489395) and serine/threonine protein kinase (gi. 16805032). To better understand their homologous entries in Cryptosporidium, further sequence similarity based BLAST searches were carried out (data not shown here).
Several hypothetical proteins of Cryptosporidium were identified from bands 2, 3 and 4 (gi.46229263, gi. 46229086, gi.32398735, gi.46229151, gi.32399022, gi.32398670). The clustered position of their constituent peptides in the respective ORF contig confirmed the existence of these hypothetical proteins in the predicted genome. Many heat shock proteins (Hsp70) were also found in a number of bands, again with high MASCOT scores for each peptide. The Hsp70 of C. parvum (gi.2894792; band 6) was found to have a maximum score (Table 1) with evidence of hits of homologous Hsp70 from other parasites such as Plasmodium, Toxoplasma and Babesia. Chaperonin protein Hsp70 has been reported in the genome of Cryptosporidium, but it could be derived from multiple intracellular locations (either from cytoplasm, ER, nucleus or mitochondria); therefore, the actual functional involvement needs further characterization.
A number of unique hits were identified in band 8, which included an EF hands domain containing protein (gi.46229170), alpha tubulin (gi.21634435), beta tubulin (gi.1944528, gi.6959876), PINT domain containing protein (gi.46229128) and mannose binding lectin type I protein (gi.46229236), while ribosomal proteins of C. parvum (gi.32398896, gi.46226494, gi.46229043, gi.32398723, gi.32399038, gi.46229021) were identified in bands 9, 15 and 16. All of these ribosomal proteins are essential structural constituents of ribosomes, while tubulins aid in GTP binding and structural molecule activity.
Cytoskeletal proteins such as actin (gi.323089) were found in more than one band (bands 1, 2, 4, 7, 8, 9), while a135 protein (gi.20513131) was identified in bands 1, 2 and 3. Actin was one of the most abundant proteins recorded during this study, being an essential part of C. parvum sporozoite motility as well as structural organization. The important glycolytic enzyme, glyceraldehyde-3-phosphate dehydrogenase (gi.46229140), was also identified from at least three different bands, supporting the hypothesis that glycolysis is the major energy source in this parasite.
The distribution of identified proteins according to theoretical molecular weight and pI demonstrate the ability of LC-MS/MS to identify proteins of high molecular mass and with extreme pH values (Fig. 3). During this experiment, it was possible to identify eight proteins (24%) with pI values > 9 and two (6%) with masses > 150 kDa. These included a protein of 282 kDa (hypothetical predicted armadillo/beta-catenin-like repeat protein with unknown function, gi.32399022) and another of 234 kDa (hypothetical protein with unknown function, gi.32399103). It is unlikely that these proteins would be identified by 2-DE analysis.
When the 33 hits of Cryptosporidium sp. identified by the MASCOT search against the NCBI database were arranged according to function (Fig. 4), 49% were found to be associated with protein biosynthesis. Other proteins involved in different functional roles included intermediate and energy metabolism (9%), cell polarity and structure (6%), protein/RNA transport (3%) and DNA maintenance (3%). A large number of hits (30%) consisted of hypothetical proteins or proteins with unknown function.
Protein expression profiles of microbial organisms have previously been produced through the combination of 1D or 2-D gel electrophoresis (2-DE) and MS. Despite being a powerful protein separation technique, 2-DE in combination with MS has a limited dynamic range and does not provide enough sensitivity to identify proteins present in low abundance in complex samples. Moreover, 2-DE has limitations at resolving membrane proteins (due to solubility) and proteins with high pH and molecular weight. Ideally, proteomics requires a comprehensive experimental approach to analysis of protein expression (i.e. qualitative and quantitative analysis of hundreds or thousands of proteins under different metabolic states). Because it is a time and labour intensive technique due to the nature of spot-by-spot analysis, 2-DE is less suitable for rapid large-scale analysis of complex protein mixtures. Therefore, any substitute for 2-DE should allow for rapid identification of proteins and deal equally well with all proteins, regardless of their abundance, subcellular localization or physicochemical parameters. The complexity of the starting material is another important issue that is essential to optimising the characterization of proteins from any biological sample. MudPIT approaches often start with highly complex samples. Prefractionation of the sample (by SDS-PAGE, liquid chromatography steps or subcellular fractionation) can help reduce the sample complexity and thus improve the resolving power of the analysis. For gel LC-MS/MS analysis, the complex sporozoite protein material was separated by 1D-SDS-PAGE prior to nano-liquid chromatography and MS. The proteins from each gel slice were digested with trypsin separately in an apparently less complex sample.
The use of 1D-SDS-PAGE to analyze the whole C. parvum sporozoite lysate was successful at separating the constituent proteins, which included both soluble and insoluble proteins. This approach has the advantage of being able to correlate apparent molecular masses with gene annotation derived from theoretical mass prediction [10]. During this study, LC-MS/MS analysis of 20 gel bands from sporozoite proteins revealed a wide range of proteins of Cryptosporidium. In total, 135 hits were recorded from LC-MS/MS analysis of all 20 gel slices, 41% of which were unique hits of Cryptosporidium. The remaining 80 (59%) protein entries were recorded as redundant hits that included repeated protein hits for which different accession numbers were assigned in the NCBI database. This occurred in cases in which the same protein was assigned using different accession numbers (for example, a single C. parvum Hsp70 protein has two different GenBank accession numbers, gi.2894792 and gi.1616783). This was demonstrated by the existence of identical peptide(s) in different hits with the same protein ID (but different accession numbers recorded in the MASCOT result page). Another important issue is that if one or more peptide(s) are common for multiple protein entities they can lead to false positive results by matching with other non-related peptides (peptides from additional proteins). Thus, although the peptides are truly present in the sample, its matched protein is actually absent. Further bioinformatic analysis using a suitable gene and protein prediction algorithm can overcome this problem.
Some of the identified peptides matched homologous proteins of closely related parasites such as Plasmodium, Toxoplasma and Eimeria sp. It can be concluded that these homologous hits from other apicomplexa represent the presence of homologous proteins in Cryptosporidium for which no accession number was available in NCBI at the time of the search. To get a better identification of these hits and optimize the use of peptide fragmentation data, sequence similarity based BLAST homology searching was performed (data not shown). After exclusion of all redundant and non-Cryptosporidium hits from the gel LC-MS/MS analysis, 33 Cryptosporidium proteins were identified. These included structural, metabolic and hypothetical proteins covering a wide range of pHs and molecular weights. Several ribosomal proteins along with other structural proteins such as actins and beta tubulins were identified after one-dimensional separation. A number of oocyst wall proteins were also present. This was due to the normal consequence of limitations in sample preparation where it was difficult to separate sporozoites from empty oocysts and partially or unexcysted oocysts. Notably, metabolic enzymes such as the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase and protein di-sulphide isomerase were also recorded. This is consistent with the hypothesis that glycolysis might be the sole energy source of C. parvum and that the parasite primarily relies on the anaerobic oxidation of glucose for energy production [1-3,5,18]. In addition, no enzymes of the TCA cycle or cytochrome respiratory pathways were revealed during this study. However, this proteome analysis is only based on the sporozoite stage of the parasite. Analysis of all life cycle stages will be important to determine the full metabolic profile of this parasite. Because there is no cell culture available for Cryptosporidium, it will be challenging to isolate different life cycle stages for their stage-specific proteome analysis. However, isolating all different developmental stages was not the objective of this study and only the partial sporozoite proteome was explored.
Complete genome sequencing of apicomplexan parasites and other microbial pathogens not only offers insight into the biology of eukaryotes, but also provides the basis for rational therapeutic strategies. The availability of 13-fold coverage of the genome sequence for the C. parvum genome [18] has given rise to a preliminary analysis of global protein expression in this parasite. It is possible that proteins that were successfully identified represent those expressed in high abundance in C. parvum sporozoites under the specific set of experimental condition used in this study. The remaining unidentified fraction of the proteome is likely to be expressed under different environmental conditions or life cycle stages, or in lower abundance. Although different developmental stages of the parasites could be analyzed as in P. falciparum [6], only the proteome of invasive sporozoite stages were investigated in this study. A similar study in Cryptosporidium was hindered by a number of factors including the size of the parasite (smaller with less protein content), poor cell culture success (limiting the source of materials), and the unavailability of suitable purification methods (for different life cycle stages). However, with the advent of new high throughput analytical techniques and equipment, as well as successful in vitro culture of Cryptosporidium sp., further stage specific analyses will provide more valuable information to understand the biology and biochemistry of Cryptosporidium.
Proteome studies of fully sequenced genomes like yeast and Plasmodium have been able to identify a significant number of proteins using published genome sequence information. In Plasmodium, a single study [6] identified 47% (2,415 out of 5,276) of the total gene product (based on the one gene for one protein concept). It is unknown how many proteins are to be expected in the proteome of C. parvum sporozoites. Based on the genome sequence of C. parvum [18], there may be as many as 4,000 gene products present in Cryptosporidium. However, only a portion of these will be expressed in the sporozoites stage, and not all will encode protein. This number is relatively small when compared to estimates for Toxoplasma (17,000 genes) [9] and Plasmodium (5,268 genes) [7].
Finally, although a number of proteins and thousands of peptides were identified in this study, it does not represent a complete analysis of the proteome of Cryptosporidium sporozoites. Nevertheless, this method clearly provides a large-scale analysis of the proteome of this organism. In addition, we have developed a successful procedure for the protein sample preparation that could be useful in future proteomic analyses of Cryptosporidium sp. The method requires further improvements and optimization, but the experimental setup in this study generated a greater number of protein identifications than combined 2-DE and MS analysis.
Figures and Tables
Fig. 1
(A) 1D-SDS-PAGE analysis of sporozoite proteins of Cryptosporidium (C.) parvum. Total no. of oocysts ~ 107. Total amount of protein~60 µg. Electrophoresed proteins were visualized with colloidal coomassie stain. Molecular weight markers (in kilodaltons) are shown on the left. (B) The lane containing C. parvum proteins was excised into 20 slices. Each gel slice was then digested by trypsin and analyzed by LC-MS/MS.
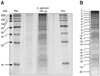
Fig. 2
Hit redundancy after LC-MS/MS analysis of different bands of 1D-SDS gel of C. parvum sporozoite protein. Redundant hits include non-Cryptosporidium hits and hits with the same identical peptides, but matching different accession numbers in the protein databases.

Fig. 3
Bimodal distribution of 33 Cryptosporidium proteins identified by a MASCOT search of LC-MS/MS data after analysing 1D-SDS-PAGE gel bands. The search was carried out against the NCBInr database. The x-axis shows the predicted isoelectric points (pI) of the proteins on a linear scale and the y-axis shows the predicted molecular weight (Mr) of the proteins on a logarithmic scale. The dotted box indicates proteins potentially amenable to analysis by 2-DE.

Fig. 4
Functional categorization of 33 Cryptosporidium proteins identified through a MASCOT search of different bands of 1D-SDS-PAGE gel. Protein categorization was made according to the MIPS functional Catalogue Database.

Table 1
Distribution of Cryptosporidium parvum hits in different bands of 1D-SDS gel*

*All proteins identified in different gel slices of 1D-SDS-PAGE gel (Fig. 1B). The peptide fragmentation data from tandem mass spectrometry (LC-MS/MS) were searched against the non-redundant NCBInr database using the MASCOT search software. The list includes all significant hits (score >35).
Acknowledgments
The author is grateful to Prof. JM Wastling of University of Liverpool for supervision of this work. Thanks also to Drs. Andy Pitt and Richard Burchmore of Sir Henry Wellcome Functional Genome Facility, University of Glasgow, Scotland, UK for their technical support with the mass spectrometry and data analysis software. The work was supported under a Commonwealth Doctoral Scholarship to the author.
References
1. Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004; 304:441–445.

2. Coombs GH. Biochemical peculiarities and drug targets in Cryptosporidium parvum: lessons from other coccidian parasites. Parasitol Today. 1999; 15:333–338.

3. Denton H, Brown SM, Roberts CW, Alexander J, McDonald V, Thong KW, Coombs GH. Comparison of the phosphofructokinase and pyruvate kinase activities of Cryptosporidium parvum, Eimeria tenella and Toxoplasma gondii. Mol Biochem Parasitol. 1996; 76:23–29.

4. Doolan DL, Southwood S, Freilich DA, Sidney J, Graber NL, Shatney L, Bebris L, Florens L, Dobano C, Witney AA, Appella E, Hoffman SL, Yates JR 3rd, Carucci DJ, Sette A. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci U S A. 2003; 100:9952–9957.

5. Entrala E, Mascaró C. Glycolytic enzyme activities in Cryptosporidium parvum oocysts. FEMS Microbiol Lett. 1997; 151:51–57.
6. Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, Witney AA, Wolters D, Wu Y, Gardner MJ, Holder AA, Sinden RE, Yates JR, Carucci DJ. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002; 419:520–526.

7. Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DMA, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002; 419:498–511.

8. Heiges M, Wang H, Robinson E, Aurrecoechea C, Gao X, Kaluskar N, Rhodes P, Wang S, He CZ, Su Y, Miller J, Kraemer E, Kissinger JC. CryptoDB: a Cryptosporidium bioinformatics resource update. Nucleic Acids Res. 2006; 34:D419–D422.
10. Lasonder E, Ishihama Y, Andersen JS, Vermunt AMW, Pain A, Sauerwein RW, Eling WMC, Hall N, Waters AP, Stunnenberg HG, Mann M. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. 2002; 419:537–542.

11. O'Donoghue PJ. Cryptosporidium and cryptosporidiosis in man and animals. Int J Parasitol. 1995; 25:139–195.
12. Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999; 20:3551–3567.

13. Sam-Yellowe TY, Florens L, Wang T, Raine JD, Carucci DJ, Sinden R, Yates JR 3rd. Proteome analysis of rhoptry-enriched fractions isolated from Plasmodium merozoites. J Proteome Res. 2004; 3:995–1001.

14. Sanderson SJ, Xia D, Prieto H, Yates J, Heiges M, Kissinger JC, Bromley E, Lal K, Sinden RE, Tomley F, Wastling JM. Determining the protein repertoire of Cryptosporidium parvum sporozoites. Proteomics. 2008; 8:1398–1414.

15. Snelling WJ, Lin Q, Moore JE, Millar BC, Tosini F, Pozio E, Dooley JSG, Lowery CJ. Proteomics analysis and protein expression during sporozoite excystation of Cryptosporidium parvum (Coccidia, Apicomplexa). Mol Cell Proteomics. 2007; 6:346–355.

16. Thompson RCA, Olson ME, Zhu G, Enomoto S, Abrahamsen MS, Hijjawi NS. Cryptosporidium and cryptosporidiosis. Adv Parasitol. 2005; 59:77–158.
17. Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Geer LY, Helmberg W, Kapustin Y, Kenton DL, Khovayko O, Lipman DJ, Madden TL, Maglott DR, Ostell J, Pruitt KD, Schuler GD, Schriml LM, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Suzek TO, Tatusov R, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006; 34:D173–D180.

18. Xu P, Widmer G, Wang Y, Ozaki LS, Alves JM, Serrano MG, Puiu D, Manque P, Akiyoshi D, Mackey AJ, Pearson WR, Dear PH, Bankier AT, Peterson DL, Abrahamsen MS, Kapur V, Tzipori S, Buck GA. The genome of Cryptosporidium hominis. Nature. 2004; 431:1107–1112.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download