Introduction
The most pressing issue in the control of parasites using anthelmintics and vaccines is rating the attractiveness of particular enzymes and metabolic pathways with respect to the likelihood of identifying specific and therapeutically useful inhibitors or potential antigen candidates. In mammals, reactive oxygen species (ROS) such as superoxide anion, hydrogen peroxide, and hydroxyl radical act as important defensive factors against parasites. Such factors damage parasite lipids, proteins, and nucleic acids, eventually causing death of the parasite. However, parasites do produce some antioxidant enzymes that cleave ROS, constituting a defense mechanism. Glutathione peroxidase (GPX) is one of the essential antioxidant enzymes in parasites [15,26]. In particular, GPXs in parasites differ from those in mammals, which contain both selenium-dependent and selenium-independent forms. Selenium-dependent GPXs play a fundamental role in protecting against ROS, whereas selenium-independent GPXs function as a backup system [15]. In contrast, the selenium-independent GPX family is the main one in parasites, with selenium-dependent GPXs basically absent [5,26]. In addition to its breaking down ROS from hosts, parasite GPXs also play roles in protecting cells against the deleterious effects of reactive oxidants generated during aerobic metabolism [9,27]. Therefore, based on their critical functions in parasite survival and metabolism, selenium-independent GPXs could be considered as potential chemotherapeutic targets and vaccine candidates for parasite control [12].
Haemonchus (H.) contortus is a blood-ingesting nematode affecting ruminants that causes major losses to the global agricultural industry every year [16,24]. Control of H. contortus has thus far been carried out using anthelmintics and grazing management. However, the excessive and uncontrolled application of chemical drugs has resulted in the emergence of anthelmintic-resistant strains of the parasite, toxic residues in the human food chain, and environmental pollution [10,25]. These undesirable effects have further led to attempts to better understand the biology of H. contortus, with the eventual goal of developing alternate or supplementary means of control, including the development of molecular vaccines. Several enzymatic antioxidants have been considered as vaccine candidates in other parasites, including superoxide dismutase (SOD) for Schistosoma (S.) mansoni, glutathione S-transferase for S. mansoni, and peroxidoxin for Fasciola hepatica [6,21]. In H. contortus, a combination of cytosolic and extracellular SODs has been tested for its potential to protect lambs from H. contortus infection, demonstrating a slight reduction in worm burdens [20].
HC29 was originally identified in a study on differential gene regulation during H. contortus development using RNA arbitrarily-primed PCR [14]. In situ analysis of adult parasites revealed expression of HC29 in all examined organs of H. contortus, particularly in the intestinal microvilli and muscle cells lining the cuticle. The previously published partial sequence of HC29 EST in H. contortus (380 bp, accession No. AF305967) possesses significant similarity with Caenorhabditis elegans GPX R03G5.5 (accession No. U51994), namely 72% identity (85% similarity) at the amino acid level over 48 residues, and therefore could be a GPX molecule. Further, another GPX of H. contortus (accession No. AY603337) was identified previously. However, protein sequence analysis indicated that it is disparated from HC29 EST [2]. Until now, the full sequence and protein characteristics of H. contortus HC29 have not been reported.
In this research, the full-length cDNA sequence of H. contortus HC29 along with the enzyme activity of the recombinant protein were evaluated.
Materials and Methods
3'-rapid amplification of cDNA ends (3'-RACE) and 5'-RACE
The 3'-end of the cDNA was amplified by a 3'-full RACE kit (TaKaRa Bio, Japan) using the gene-specific primers 3 outer primer (OUP) and 3 inner primer (INP) (Table 1), which were designed based on H. contortus EST (GenBank accession No. AF305967) in combination with the 3'OUP and 3'INP in the kit (Table 1).
The 5-end of the cDNA was amplified by 5'-RACE PCR using a 5'-full RACE kit (TaKaRa Bio, Japan). Primary PCR was performed using the primers 5OUP (Table 1) and 5'OUP, followed by a second PCR using 5INP (Table 1) and 5'INP.
Products from both of the second-round PCRs were recovered using an agarose gel DNA purification kit (ver. 2.0; TaKaRa Bio, Japan) according to the manufacturer's instructions and then ligated into pMD-18T cloning vector (TaKaRa Bio, Japan). Clones containing inserts of the expected size were identified by XbaI and HindIII digestion and then sequenced by Invitrogen, USA. The complete sequence of the cDNA was deduced from the overlapping sequences of both amplification products using BioEdit (ver. 7.0.1; North Carolina State University, USA).
Synthesis of complete HC29 cDNA and open reading frame
The full-length cDNA and open reading frame (ORF) of HC29 cDNA were generated by RT-PCR using the primers QCS and QCA for the full-length cDNA and ORFS and ORFA for ORF (Table 1). The purified PCR products were ligated into pMD-18T cloning vector (TaKaRa Bio, Japan), transformed, and then sub-cultured. Randomly selected clones containing inserts of the expected size were then identified by XbaI and HindIII digestion. The sequences of the clones were also verified by Invitrogen (USA). All oligonucleotides used in this study (Table 1) were synthesized by Invitrogen (USA).
Sequence analysis
BLASTP and BLASTX were used for sequence similarity searches between HC29 and reported GPX [1]. The sequences of both HC29 and the reported GPX protein were aligned using Clustal W 1.82 [33]. Protein motifs, glycosylation sites, and secondary structures were predicted using programs accessible on the Internet, including Motifscan [11] and PSIpred [22]. Phylogenetic tree analysis was generated using Clustal X 2.0 [19] and MEGA 4.1 [30].
Construction of expression vector
Expression vector of recombinant HC29 protein was constructed by inserting full-length HC29 ORF into pET-28a plasmid (TaKaRa Bio, Japan). Briefly, recombinant pMD-18T containing HC29 ORF was digested with EcoRI and XhoI, after which the HC29 ORF fragment was inserted into pET-28a vector (TaKaRa Bio, Japan) that was previously linearized with similar enzymes to give pET-28a/HC29 expression vector. The recombinant vector was then transformed into Escherichia (E.) coli DE3 stain competent cells, and positive clones were verified by enzyme digestion.
Expression of recombinant HC29 protein
Transformed E. coli harboring pET-28a/HC29 was sub-cultured in Luria Bertani media supplemented with kanamycin (100 µg/mL) and incubated at 37℃ until an OD600 of 0.4~0.6. Expression was induced with isopropyl-β-D-thiogalactopyranoside (Sigma, USA) to a final concentration of 1 mM. After 5 h of incubation at 37℃, bacteria cells were harvested and expression assayed by SDS-PAGE.
Purification of recombinant HC29 proteins
Following induction, bacterial pellets were collected, after which recombinant proteins were harvested by centrifugation, lysed for 30 min on ice in PBS buffer, and sonicated. After centrifugation at 10,000 × g, the supernatant was loaded onto a Ni2+-nitrilotriacetic acid column (GE Healthcare, USA) and purified according to the manufacturer's instructions. An elution buffer (40 mM NaH2PO4, 300 mM NaCl, pH 8.0) containing various amounts of imidazole was then evaluated for optimal release of the His-tagged proteins from the nickel resin. Purity of the proteins was estimated by 12% SDS-PAGE, and protein concentration was determined by Bradford assay [4] using bovine serum albumin as a standard.
Generation of polyclonal antibodies
To prepare polyclonal antibodies, about 0.3 mg of the purified HC29 protein was mixed with Freund's complete adjuvant, which was injected subcutaneously into SD rats (Experimental Animal Center of Jiangsu, China) at multiple places. Rats were then boosted four times at 2-week intervals after the first injection, and bleeding was carried out 10 days after. The serum containing specific anti-HC29 polyclonal antibodies was then collected, sampled, and stored at -20℃. Goat sera used in this research were collected from naturally infected goats.
Western blot analysis
Western blotting was performed after 12% SDS-PAGE. Separated protein bands, whole protein of adult worms, and purified recombinant protein were then electrophoretically transferred from the gel slab to a nitrocellulose filter (Sigma, USA) using a Trans-Blot system (Bio-Rad, USA). This was followed by incubation for 1 h in blocking solution containing 5% (w/v) skimmed dry milk, 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.01% (w/v) NaN3, and 0.05% (v/v) Tween-20, followed by incubation with primary antibodies (rat sera and naturally infected goat sera) for 1 h at 37℃ (rat sera diluted 1 : 1,000 in 5% skimmed dry milk powder/TBS and goat sera diluted 1 : 100). Secondary antibody (HRP-conjugated goat anti-rat IgG, HRP-conjugated rabbit anti-goat IgG; Sigma-Aldrich, USA) diluted 1 : 5,000 in 5% dry milk powder/TBST (TBS in 0.05% Tween-20) was incubated through a nitrocellulose filter for 1 h at 37℃ one more time. The immunoreaction was visualized using freshly prepared DAB (Sigma-Aldrich, USA) as a chromogenic substrate after 2~5 min.
Determination of recombinant HC29 protein enzymatic activity
Enzymatic activity was investigated by DTNB assay as previously described [13]. In the DTNB assay, these enzymes catalyzed the reduction of H2O2 by oxidizing glutathione (GSH), and GPX activity was confirmed by the deduction of GSH. The enzyme tubes were incubated at 37℃ containing 1 mL of 2 mM GSH, 1 mL of 0.4 M sodium phosphate buffer (pH 7.0) with 4 × 10-4 M EDTA, 0.5 mL of 0.01 M NaN3 (to inhibit catalase), 100 µg of recombinant HC29 protein, and water to a total volume of 4 mL. After 5 min of preincubation, 1 mL of 1.25 mM H2O2 (prewarmed to 37℃) was added. Following this, at 3 min intervals, 1 mL aliquots of the incubation mixture were added to 4 mL of metaphosphoric acid precipitation solution as the filtrate. GSH concentration was then determined by mixing 2 mL of filtrate with 2 mL of 0.4 M Na2HPO4 and 1 mL of DTNB reagent. A412 was recorded within 2 min after mixing. An enzyme-free tube (with H2O substituted for enzyme) acting as a negative control was established simultaneously. HC29 group and enzyme-free group were carried out with five replications per group.
Results
Characteristics of HC29 cDNA sequence
The product of 5'-RACE-PCR was a fragment of 523 bp. A 22 bp nematode-specific spliced leader sequence type 1 (SL1) was identified from 5 to 26 bp at the 5'-end of this fragment. The 3'-RACE-PCR product contained a polyadenylation tail at position 498 bp downstream of stop codon TAG. HC29 transcript was 1,113 bp long and obtained by splicing both the 3'- and 5'-RACE fragments. Its nucleotide and inferred amino acid sequences were submitted to GenBank under accession No. GQ927327. The sequence AATGAA at position 18 bp upstream of the polyadenylation site was identified as a distal polyadenylation stop signal of the HC29 cDNA gene. The ORF from 65 to 571 bp was 507 bp in size (Fig. 1) and encoded a polypeptide of 168 amino acid.
Characteristics of amino acid sequence of HC29
The predicted protein of HC29 consisted of 58 hydrophobic amino acids, 43 hydropolar amino acids, 24 strongly basic amino acids, and 23 strongly acidic amino acids. A search of the protein databases using the Basic Local Alignment Search Tool network service (National Center for Biotechnology Information, USA) indicated that the encoded protein had 44.7 to 80.4% similarity with a number of GPXs in the thioredoxin-like family (Table 2). The phylogenetic tree generated from the protein sequences (Fig. 2) revealed a close relationship between HC29 and GPX of Caenorhabditis elegans.
Amino acid sequence alignment between HC29 and the GPXs of other species revealed that HC29 protein possessed the catalytic triad active site residues C38, Q73, and W127 (Fig. 3). The dimer interface sites G72, P75, C77, D80, N83, and N87 were also predicted in the GPXs, indicating a tetrameric structure for HC29. A GPX (GKvLIIvNVaSqCGlT) was identified in the putative amino acid sequence at position 26~41 by the ScanProsite database tool. Two N-glycosylation sites (44NYTQ47, 128NLTK131) were predicted by the CBS web server (Center for Biological Sequence Analysis, Denmark).
Alignment of HC29 and previous GPX from H. contortus
DNA sequence alignment between HC29 and H. contortus GPX showed that both sequences possessed different types of SL sequences (Fig. 4). HC29 had a SL1 type sequence (5GGTTTAATTACCCAAGTTTGAG26), whereas GPX had a SL2 type sequence (1GGTTTTAACCCAGTTACTCAAG22), as reported by Bagnall and Kotze [2].
Alignment of the amino acid sequence of HC29 with that of reported GPX revealed 30.8% identity and 48.6% similarity at the amino acid level (Fig. 5). A cysteine residue coded by UGU located in the active site is a specific structure of selenium-independent GPXs, whereas selenium-dependent GPXs possess a seleno-cysteine residue coded by UGA. Both HC29 and the reported GPX protein contained a cysteine residue coded by UGU in their active sites (position 38 for HC29; position 50 for GPX). The amino acid sequence of HC29 possessed the same catalytic triad active site residues C38, Q73, and W127 as those of the reported GPX. However, the predicted dimer interface site residues of HC29 were different. Specifically, the dimer interface site residues of HC29 consisted of G72, P75, C77, D80, N83, and N87, whereas those of the reported GPX consisted of L83, P86, E88, E91, N94, and Y98. Furthermore, HC29 contained a GPX motif sequence (GKvLIIvNVaSqCGlT) at amino acid position 26~41, whereas the reported GPX had a GPX motif sequence (GQvLLIiNVaTfCAyT) at amino acid position 38~53.
Expression of recombinant HC29 protein
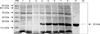
The expression product from E. coli migrated to an approximate molecular mass of 22 kDa, as visualized under reducing conditions (Fig. 6, Lanes 4~8). Both the E. coli transformed with pET-28a and the cells transformed with recombinant plasmid before induction did not produce expressive target bands (Fig. 6, Lanes 1~3).
Western blot assay
Native protein extracts of H. contortus probed with rat anti-recombinant HC29 serum displayed a single band with a size nearly the same as that of the recombinant protein (Fig. 6).
There was no detectable band in the Western blot analysis of recombinant HC29 protein, which was electro-transferred to a nitrocellulose membrane and probed with serum from goats naturally infected with H. contortus parasites as the primary antibody (data not shown).
Discussion
A number of GPXs were isolated or recombined from a wide range of nematodes [15]. However, previous reports on GPX from H. contortus are quite limited in number. In the present investigation, the complete sequence of a H. contortus GPX cDNA termed HC29 was cloned and characterized for the first time. In a comparison with the predicted amino acid sequences of other GPXs, HC29 demonstrated 44.7~80.4% similarity with GPX homologues in thioredoxin-like family. The conserved motif of the catalytic register along with the conserved catalytic triad active site residues C38, Q73, and W127 of thioredoxin-like family GPXs were identified in HC29 protein. Enzymatic activity assay of recombinant HC29 protein showed that it could catalyze H2O2 by oxidizing GSH to oxidized glutathione. All of the above results suggest that HC29 is a GPX of the thioredoxin-like family. GPXs in mammals include both selenium-dependent and selenium-independent enzymes, the differences between which lie in the catalytic triad active sites and 'stem-loop'. In selenium-independent GPXs, the active sites consist of C-Q-W. However, in selenium-dependent ones, the cysteine is substituted by seleno-cysteine. On the other hand, the 'stem-loop' is a specific RNA structure present only in the 3'-untranslated region (UTR) of selenium-dependent GPXs and functions in recognizing UGA as a seleno-cysteine residue and not a stop codon [28]. In this research, we observed a cysteine residue at position 38 as well as the lack of a 'stem-loop' sequence in the 3'-UTR by using RNA draw software to analyze the full RNA sequence. Based on our results, we could conclude that HC29 is a selenium-independent enzyme.
In a DNA sequence comparison, HC29 was found to be different from the reported GPX of H. contortus (accession No. AY603337) [2]. DNA sequence comparison also revealed that the reported GPX possessed SL2, whereas HC29 possessed SL1. Amino acid sequence alignment between HC29 and the reported GPX revealed that these two shared 30.8% identity and 48.6% similarity, along with different dimer interface sites. Furthermore, HC29 displayed a GPX motif sequence (GKvLIIvNVaSqCGlT) at position 26~41, whereas the reported GPX had a different GPX motif sequence (GQvLLIiNVaTfCAyT) at position 38~53. These results indicate that the HC29 is a novel GPX of H. contortus.
Most nematode mRNAs possess a 5'-SL sequence. SL sequences in nematodes are generated by trans-splicing, which is vital to the maturation of pre-mRNAs. SL1 was the first SL ever reported in nematodes, and a series of SL sequences have been subsequently described [3]. SL1 plays main roles in regulating the optimal length of the 5'-UTR in mature mRNA as well as the formation of nucleotide construction benefit for gene translation [29,35]. However, SL2 is closely linked (temporally or mechanistically) to 3'-end formation and polyadenylation of upstream genes [17]. Further, the sequence of SL2 is quite distinct from that of SL1. Genes containing SL2 all appear to be located within ~150 bases downstream of another gene containing SL1 on the same strand [18]. Therefore, the reported GPX was determined to be consecutively downstream of HC29 on the same strand of the primary transcript.
A dimer interface plays an important role in the formation of enzyme dimers. Existence of dimer interface sites in GPXs indicates that these enzymes possess a tetrameric structure, or a dimer of dimers. Both HC29 and the reported GPX of H. contortus were determined to possess dimer interface sites, which implies a polymeric structure.
Comparison with the amino acid sequence of the reported GPX showed that HC29 also possessed the same catalytic triad active sites C-Q-W, suggesting that both GPXs belong to the GPX superfamily. Analysis of the GPX motif sequence using the ScanProsite database showed that HC29 had a GKvLIIvNVaSqCGlT motif, whereas the reported GPX had the sequence GQvLLIiNVaTfCAyT. Although these motif sequences are different, both are conserved in the GLUTATHIONE_PEROXID_1 (accession No. Ps00460, Prosite) family. Therefore, we could conclude that HC29 and the reported GPX of H. contortus belong to the GLUTATHIONE_ PEROXID_1 (accession No. Ps00460, Prosite) family.
The amino acid sequence comparison also showed that both HC29 and the reported GPX possessed the same cysteine residue in their active sites and both were selenium-independent GPXs. The cysteine residues were found to be highly conserved in the GPXs of Brugia pahangi, Brugia malayi, Wuchereria bancrofti, and Dirofilaria immitis [8,34]. Therefore, these findings suggest that the GPXs of nematode are selenium-independent.
It has been reported that, in all aerobic organisms, selenium-dependent GPXs are the main enzymes for scavenging H2O2 and preventing the further formation of ROS. In addition, selenium-independent GPXs have been found to adequately function as a back-up system for selenium-dependent GPXs in the absence of sufficient concentrations of selenium [15]. Surprisingly, these selenium-dependent enzymes are absent in parasitic nematodes, including H. contortus and filarial nematodes. This lack of enzymes capable of dealing with H2O2 is puzzling and should be further researched [5,26].
In this research, low but significant enzymatic activity was observed in recombinant HC29 protein by Hafeman assay [13]. Similarly, Tang et al. [31] previously found that a recombinant selenium-independent GPX of Brugia phangi expressed in insect cells (gp29) shows very low activity against H2O2. This indicated that the recombinant GPX of nematode has low enzyme activity under artificial conditions or that H2O2 is not the optimal substrate of this enzyme. To address this, Tang et al. [32] suggested that GPXs mainly play roles in repairing oxidatively damaged membranes rather than acting directly as antioxidant enzymes. This could explain the low activity of the recombinant GPX under artificial conditions. However, exactly why recombinant GPX of nematode displays low activity under artificial conditions should be further researched. In this study, the immunoblot assay results indicated that the native HC29 of H. contortus was 22 kDa in size, similar to that of the recombinant protein. This suggests that perhaps GPX does not undergo post-translational modification or only a small modification occurs that is undetectable by SDS-PAGE.
HC29 was expressed particularly in the intestinal microvilli and muscle cells lining the cuticle [14]. However, in this research, the recombinant protein was not recognized using naturally infected serum. This suggests that the native HC29 of H. contortus is neither secreted into its native host tissues under physiological conditions nor recognized by the host immune system. As this result runs contrary to the known location of HC29, more investigations are needed.




 PDF
PDF ePub
ePub Citation
Citation Print
Print









 XML Download
XML Download