Abstract
Campylobacter (C.) fetus (epsilonproteobacteria) is an important veterinary pathogen. This species is currently divided into C. fetus subspecies (subsp.) fetus (Cff) and C. fetus subsp. venerealis (Cfv). Cfv is the causative agent of bovine genital Campylobacteriosis, an infectious disease that leads to severe reproductive problems in cattle worldwide. Cff is a more general pathogen that causes reproductive problems mainly in sheep although cattle can also be affected. Here we describe a multiplex PCR method to detect C. fetus and differentiate between subspecies in a single step. The assay was standardized using cultured strains and successfully used to analyze the abomasal liquid of aborted bovine fetuses without any pre-enrichment step. Results of our assay were completely consistent with those of traditional bacteriological diagnostic methods. Furthermore, the multiplex PCR technique we developed may be easily adopted by any molecular diagnostic laboratory as a complementary tool for detecting C. fetus subspecies and obtaining epidemiological information about abortion events in cattle.
Members of the genus Campylobacter are Gram-negative epsilonproteobacteria highly adapted to vertebrate hosts. Most Campylobacter species are pathogens in a wide range of livestock species [18] and also have extensive reservoirs in wildlife [3]. In particular, the species Campylobacter (C.) fetus is an important veterinary pathogen that can also infect humans. This species is currently divided into C. fetus subspecies (subsp.) fetus (Cff) and C. fetus subsp. venerealis (Cfv). A phenotypic variant designated as C. fetus subsp. venerealis biovar intermedius (Cfvi) has also been described [24].
Cff causes abortion mainly in sheep [6] and to a lesser extent in cattle. In humans, this is an opportunistic microorganism that mainly infects immune-compromised patients [12,21]. Cfv is the causative agent of bovine genital campylobacteriosis (BGC), an infectious venereal disease which leads to reproductive problems such as infertility, lowered pregnancy rates, and abortion [4]. This subspecies is thought to be a cattle-specific pathogen and has been isolated from bull preputial cavities, cow vaginal mucus, and the organs of aborted fetuses [16,18]. Although Cfvi shows characteristics similar to those of Cfv, it is unclear whether it can occur only in the genital tract or both the genital and intestinal tracts [5,6]. Campylobacteriosis is a matter of concern for the cattle industry worldwide as this disease causes significant economic losses [2,15]. Infected animals are subjected to trade restrictions and it is therefore mandatory to report outbreaks to the World Organization for Animal Health (OIE).
In order to collect epidemiological information and other clinically relevant data about C. fetus infection, adequate diagnostic methods are required. Bacteriological analyses, such as culture isolation and biochemical tests, are useful for evaluating different kinds of samples even with low bacterial counts (e.g., preputial washings from bulls). Although these methods are well standardized and extensively used, they are laborious and time-consuming, thus making them disadvantageous when processing large-scale samples or delivering a fast diagnosis. Molecular methods using polymerase chain reaction (PCR) have become a good alternative for obtaining rapid and highly specific bacterial diagnoses [8,11,27]. There are relatively few assays described for C. fetus [1,10,23,25]. The most widely used is a multiplex PCR technique that detects C. fetus and differentiates between its subspecies [7]. This assay amplifies a 764-bp fragment that is specific for C. fetus, and an additional 142-bp fragment that appears in Cfv but not Cff [7]. However, the amplicon used as a marker for Cfv has been reported to produce results that contradict those of other assays [7,19]. New markers for Cfv have been suggested [1,23], but have not been extensively applied.
In the present study, we developed a multiplex PCR assay to amplify a C. fetus-specific 764-bp sequence (C. fetus marker) and a 233-bp sequence that is only present in Cfv (Cfv marker). The latter marker was recently identified by whole-sequence genome comparisons between both subspecies [13]. This multiplex PCR assay allowed straightforward detection of the species C. fetus and Cfv subspecies, and was standardized using cultured strains. The assay was also successfully used to evaluate DNA extracted directly from the abomasal liquid contents of aborted bovine fetuses without any pre-enrichment step.
The multiplex PCR assay included two sets of commercial primers synthesized by Integrated DNA Technologies (USA). These primer sets were previously described in the literature but had never been used simultaneously (Table 1). The primer set MG3F/MG4F amplifies a 764-bp region of the cstA gene, which is a widely used genetic marker for C. fetus [7]. The primer set nC1165g4F/nC1165g4R amplifies a 233-bp amplicon of the virB11 gene that is present in Cfv but not in Cff [13]. Multiplex PCR using these two primer sets would accordingly produce only the 764-bp amplicon for Cff, and yield both the 764-bp and 233-bp amplicons in the case of Cfv.
Standardization of the multiplex PCR assay was performed with cultured Uruguayan, Australian, and Argentinian strains of Cff, Cfv, and Cfvi, provided from reference laboratories of Division of Veterinary Laboratories (DILAVE), Uruguay and National Institute of Agricultural Technology (INTA), Argentina, that are routinely used as controls for diagnostic procedures in Uruguay. Abomasal liquid from an Uruguayan aborted fetus that had been stored at -20℃ since 1998 was included as an additional control. A bacterial strain characterized as Cff had been previously isolated from this liquid in DILAVE. The multiplex-PCR assay was also performed with 10 Uruguayan samples collected from the field during 2010. These samples were directly obtained from the abomasal contents of bovine aborted fetuses (Table 2). C. jejuni, C. sputorum subsp. bubulus (Csb), C. coli, C. hyointestinalis, and Escherichia (E.) coli were used as negative controls (INTA, Argentina) (Table 2); these are Uruguayan and Argentinian strains that are routinely used as negative controls for diagnostic laboratories.
Control and field strains were previously typed using standard bacteriological methods for Campylobacter fetus subespecies differentiation [9] in order to test the multiplex-PCR assay specificity. Bacterial samples were grown in heart brain infusion agar (Sigma-Aldrich, USA) and Campylobacter selective supplement (Thermo Fisher Scientific, USA) as selective medium under microaerophillic conditions for 48 h at 37℃. The presumptive Campylobacter colonies were grown in Brucella broth (Sigma-Aldrich, USA) with 1% glycine (Sigma-Aldrich, USA) and in Brucella broth with NaCl and cysteine (Sigma-Aldrich, USA) to detect H2S production with lead acetate paper (Sigma-Aldrich, USA).
DNA was extracted from 500 µL of control strain colonies (1 × 108 CFU/mL) resuspended and diluted 10-fold in PBS (pH 7.4) using a QIAamp DNA Mini Kit (Qiagen, Germany) according to the manufacturer's protocol. The DNA purification protocol was the same for the ten field samples although 1,000 µL of liquid abomasal content from the aborted fetuses was used. Nanodrop 1000 (Thermo Scientific, USA) was used to assess the extracted DNA purity, measured as the ratio of absorbance at 260 and 280 nm (A260/280).
The multiplex PCR reaction was first optimized with different concentrations of specific components, including MgCl2, Taq, and dNTPs, as well as various ratios of both primer sets. The best results were obtained when 1 µL of DNA was used in a 20-µL reaction containing 0.5 mM of each dNTP (Thermo Scientific, USA), 2 µL of 1× reaction buffer (Thermo Scientific, USA), 2.5 mM MgCl2 (Thermo Scientific, USA), 0.625 µM MG3F/MG4R primer set, 0.375 µM nC1165g4F/nC1165g4R primer set, and 1.5 U Taq DNA polymerase (Thermo Scientific, USA). A Corbett PCR Thermal Cycler (Qiagen, Germany) was used for amplification. The following cycling conditions were used: an initial denaturation for 3 min at 95℃ followed by 35 cycles of denaturation for 30 sec at 94℃, annealing for 30 sec at 53℃, and extension for 1 min at 72℃. Amplicons were separated in 1.0% agarose gels and stained with ethidium bromide.
Amplicons from one of the field samples (6600) were purified using a QIAquick PCR Purification Kit (Qiagen, Germany) and sequenced using an ABI Prism 377 automated sequencer (Applied Biosystems, USA). The resulting sequences were submitted to the GenBank database, and the Basic Local Alignment Search Tool (National Center for Biotechnology Information, USA) was used to measure their similarity with publicly available DNA sequences.
To measure assay sensitivity, two different approaches were used. First, five serial 2-fold dilutions of abomasal liquid from field samples known to be positive were prepared and DNA was isolated. DNA quantification was then performed using a Nanodrop 2000 (Thermo Scientific, USA). Dilutions containing 40, 20, 10, 5, and 2.5 ng of DNA/µL were thus made. One µL of purified DNA from each dilution was used for the PCR reactions.
Second, known dilutions (101, 103, 105, and 107 CFU/mL) of Cff and Cfv were mixed with 1 mL of bull semen extracted from commercial insemination syringes [20] or macerated cattle lungs (INTA, Argentina). Both of these materials were originally negative for Campylobacter. DNA was extracted from each mixture using a QIAamp DNA Mini Kit (Qiagen, Germany) following the manufacturer's protocol for tissues. Multiplex PCR assay was performed using 1 µL of each DNA sample.
C. fetus control strains were tested using standard bacteriological methods. Cff063, CffA28, CffC1N3, Cff584, and Cff710 grew in 1% glycine and produced H2S. Accordingly, they were classified as Cff. On the other hand, Cfv3598, CfvD78, Cfv608, and Cfv371 did not grow in 1% glycine and were unable to generate H2S. This resulted in their designation as Cfv. Cfvi470 did not grow in 1% glycine but produced H2S; this microorganism was thus classified as Cfvi (Table 2).
None of the field strains were able to grow in 1% glycine or produce H2S. Therefore, all were classified as Cfv (Table 2).
The multiplex PCR assay was standardized using genomic DNA extracted from previously cultured control strains. No PCR products were recovered with template DNA from non-C. fetus bacterial species that were used as negative controls (Table 2). PCR reactions with template DNA from Cfv and Cfvi yielded 764-bp and 233-bp amplicons. When using template DNA from Cff, only the species-specific 764-bp amplicon was produced (Table 2 and Fig. 1). Non-specific amplification was not observed at any time.
Once standardized with the control strains, the multiplex PCR assay was conducted to analyze DNA extracted from the 10 field samples without any pre-enrichment steps. Initial identification of the strains present in the field samples (Cfv) was confirmed by the production of two amplicons with the expected sizes (Table 2 and Fig. 2).
Specificity of the multiplex PCR, assessed by comparison with phenotypic data, was further confirmed by amplicon sequencing. The sequence of the C. fetus amplicon (GenBank accession No. JF893945) was 100% identical to the cstA gene in the reference Cff 82-40 genome (GenBank accession No. NC_008599) and with three other published sequences of this gene (GenBank accession Nos. AY158813, AY158814, and NZ_ACLG01001165). The Cfv amplicon (GenBank accession No. JF901335) was 100% identical to the virB11 gene in the Cfv Azul-94 genome (GenBank accession No. NZ_ACLG01001023).
Sensitivity of the multiplex PCR was measured using decreasing dilutions of DNA obtained from the field samples. Assay sensitivity essentially remained unchanged when using 40 to 5 ng/µL of template DNA from the abomasal liquid. No PCR products were produced when using 2.5 ng/µL of the template DNA. All DNA dilutions had an A260/280 ratio between 1.8 and 2.0, indicating relatively high purity. Results of this study indicated that the detection limit for DNA from abomasal liquid is between 5 and 2.5 ng/µL.
Sensitivity was further analyzed by examining semen and lung samples spiked with known concentrations of bacteria. PCR amplification was observed when using 1 µL of sample from semen or lung inoculated with 103, 105, and 107 CFU/mL but not 101 CFU/mL. This finding indicates that the detection limit for bacteria in a sample is around 103 CFU regardless of the source material (semen or lung).
The multiplex PCR assay described herein is able to identify the species C. fetus and distinguish between its subspecies in a single step. The assay design is based on PCR amplification of a species-specific marker to identify C. fetus and an exclusive Cfv sequence that discriminates between subspecies. The species-specific marker has been widely used for the molecular diagnosis of C. fetus and no cross-reaction has been reported with other bacterial species [7,23]. This marker was used in the only multiplex PCR assay previously described in the literature for identifying species and subspecies of C. fetus. However, the subspecies marker originally included in this method showed cross-reactivity with Cff and other Campylobacter species [7,19]. Although other markers for subspecies differentiation have been proposed [1,23], the most significant progress in this field has come from whole genome comparison between subspecies; this comparison identified a pathogenicity island present in Cfv but not Cff which contains many genes that could be potentially used as subspecies-specific markers for Cfv [13] . In the current investigation, we selected a 233-bp sequence present in the Cfv pathogenicity island to perform subspecies differentiation with our multiplex PCR assay. Under optimal conditions, this exclusive Cfv sequence was amplified with the same efficiency as the C. fetus species-specific marker in the PCR reaction.
The assay was standardized using cultured control strains that allowed the rapid, straightforward detection of C. fetus and subspecies discrimination based on the presence or absence of specific amplicons. Cff was identified by the presence of only one amplicon (764 bp) while Cfv resulted in two amplicons (764 bp and 233 bp) of distinct sizes. This multiplex PCR assay produced the same amplicon pattern for Cfvi and the other Cfv strains we tested. However, Moolhuijzen et al. [13] analyzed one strain of Cfvi using the same primer set and found that this subspecies had the same profile as Cff. To date, a specific set of primers that can distinguish Cfvi from other C. fetus strains has not been validated, suggesting that this subspecies represents a heterogeneous subset within C. fetus [13]. A more definitive genetic characterization of this biovar should be performed, particularly as it seems to be relatively common in some adult cattle populations [16].
Application of the assay for analyzing cultured samples can be used as a complement to biochemical methods since our PCR-based technique reduces the diagnosis time by avoiding the need for additional tests to identify subspecies. It is noteworthy that this multiplex PCR assay can be used for samples taken directly from aborted fetuses. To the best of our knowledge, only one recent report exists in which PCR methods were used for analyzing DNA directly extracted from abomasal liquid to detect C. fetus [22]. However, our report is the first that describes subspecies discrimination using abomasal liquid as starting material.
The methodology we developed represents a good tool for obtaining epidemiological information, like frequency and etiology, pertaining to bovine abortions. A main advantage of this assay is that the whole process (DNA extraction, multiplex PCR, and gel electrophoresis) allows the detection and discrimination of C. fetus subspecies in approximately 5 h in contrast to traditional bacteriological methods that require about 1 week. Nucleotide sequences of the amplicons obtained from aborted fetuses by multiplex PCR were identical to those found in the GenBank database, so these markers seemed to be conserved and appropriate for diagnostic purposes. The assay also produced negative results for C. jejuni, Csb, C. coli, C. hyointestinalis, and E. coli, demonstrating high specificity even with closely related species. These findings demonstrated that the assay was correctly standardized and proved to be sensitive when using as little as 5 ng of DNA extracted from the abomasal content of aborted bovine fetuses. Adequate sensitivity was also achieved using tissues and body fluids inoculated with at least 103 CFU. Hence, it is feasible to perform standardization using other kinds of non-cultured samples, like preputial washings or vaginal mucus, with low bacterial counts.
It is well known that Cfv is responsible for chronic infections leading to continuous reproduction problems although the association of this subspecies with abortion is not clear. We reviewed publications [1,14,17,26] in which aborted fetuses were used as samples and found that 41 cases of abortion (76%) were due to Cfv while 13 (24%) were caused by Cff. Accordingly, Cfv caused the vast majority of abortions in cattle compared to Cff that is widely recognized for its association with sheep abortion [6]. The 10 field samples analyzed in the current study were obtained in 2010 and all were found to exclusively contain Cfv. These results are indicative of Cfv being the most relevant subspecies involved in abortion events in Uruguayan cattle.
Although some variability has been reported for traditional bacteriological methods (growth in 1% glycine and H2S production) used to differentiate C. fetus subspecies, these techniques are extensively regarded as the gold standard. When comparing data from these traditional methodologies, our multiplex PCR assay was able to correctly identify subspecies in all cases. This finding, together with the easy and quick performance of our PCR-based technique, supports the potential use of our method not only as a routine diagnostic test for BGC but also as a high throughput method for obtaining relevant epidemiological data.
Figures and Tables
 | Fig. 1Multiplex PCR results for C. fetus subsp. fetus (Cff), C. fetus subsp. venerealis (Cfv), C. fetus subsp. venerealis biovar intermedius (Cfvi), and the negative control strains. Lane 1: negative control (no template), Lane 2: 200-bp molecular weight ladder, Lane 3: Cff063, Lane 4: CffA28, Lane 5: Cfv3598, Lane 6: CfvD78, Lane 7: Cfvi470, Lane 8: C. sputorum subsp. bubulus, Lane 9: C. jejuni, Lane 10: C. hyointestinalis, Lane 11: C. Coli, and Lane 12: E. coli. |
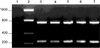 | Fig. 2Multiplex PCR results of the four field samples. Lane 1: negative control (no template), Lane 2: 200-bp molecular weight ladder, Lane 3: Cfv positive control, Lane 4: 6600, Lane 5: 3726, Lane 6: 3837, and Lane 7: 2733. The four field samples were identified as Cfv. |
References
1. Abril C, Vilei EM, Brodard I, Burnens A, Frey J, Miserez R. Discovery of insertion element ISCfe1: a new tool for Campylobacter fetus subspecies differentiation. Clin Microbiol Infect. 2007. 13:993–1000.

2. Campero CM, Anderson ML, Walker RL, Blanchard PC, Barbano L, Chiu P, Martínez A, Combessies G, Bardon JC, Cordeviola J. Immunohistochemical identification of Campylobacter fetus in natural cases of bovine and ovine abortions. J Vet Med B Infect Dis Vet Public Health. 2005. 52:138–141.

3. Debruyne L, Gevers D, Vandamme P. Nachamkin I, Szymanski CM, Blaser MJ, editors. Taxonomy of the family Campylobacteraceae. Campylobacter. 2008. 3rd ed. Washington: ASM Press;3–25.
4. Dekeyser J. Butzler JP, editor. Bovine genital campylobacteriosis. Campylobacter Infection in Man and Animals. 1984. Boca Raton: CRC Press;181–191.

5. Elazhary M. An assay of isolation and identification of some animal vibrios and of elucidation of their pathological significance. Meded Veeartsschool Rijksuniv Gent. 1968. 12:1–80.
6. Garcia MM, Eaglesome MD, Rigby C. Campylobacters important in veterinary medicine. Vet Bull. 1983. 53:793–818.
7. Hum S, Quinn K, Brunner J, On SLW. Evaluation of a PCR assay for identification and differentiation of Campylobacter fetus subspecies. Aust Vet J. 1997. 75:827–831.

9. Lecce JG. Some biochemical characteristics of Vibrio fetus and other related Vibrios isolated from animals. J Bacteriol. 1958. 76:312–316.

10. Linton D, Owen RJ, Stanley J. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res Microbiol. 1996. 147:707–718.

11. Malorny B, Tassios PT, Rådström P, Cook N, Wagner M, Hoorfar J. Standardization of diagnostic PCR for the detection of foodborne pathogens. Int J Food Microbiol. 2003. 83:39–48.

12. Meier PA, Dooley DP, Jorgensen JH, Sanders CC, Huang WM, Patterson JE. Development of quinolone-resistant Campylobacter fetus bacteremia in human immunodeficiency virus-infected patients. J Infect Dis. 1998. 177:951–954.

13. Moolhuijzen PM, Lew-Tabor AE, Wlodek BM, Agüero FG, Comerci DJ, Ugalde RA, Sanchez DO, Appels R, Bellgard M. Genomic analysis of Campylobacter fetus subspecies: identification of candidate virulence determinants and diagnostic assay targets. BMC Microbiol. 2009. 9:86.

14. On SLW, Harrington CS. Evaluation of numerical analysis of PFGE-DNA profiles for differentiating Campylobacter fetus subspecies by comparison with phenotypic, PCR and 16S rDNA sequencing methods. J Appl Microbiol. 2001. 90:285–293.

15. Repisso MV, Gil A, Bañales P, D'Anatro N, Fernandez L, Guarino H, Herrera B, Núñez A, Olivera M, Osawa T, Silva M. Prevalencias de las principales enfermedades infecciosas que afectan el comportamiento reproductivo en la ganadería de carne y caracterización de los establecimientos de cría del Uruguay. Veterinaria. 2005. 40:5–28.
16. Schmidt T, Venter EH, Picard JA. Evaluation of PCR assays for the detection of Campylobacter fetus in bovine preputial scrapings and the identification of subspecies in South African field isolates. J S Afr Vet Assoc. 2010. 81:87–92.

17. Schulze F, Bagon A, Müller W, Hotzel H. Identification of Campylobacter fetus subspecies by phenotypic differentiation and PCR. J Clin Microbiol. 2006. 44:2019–2024.

18. Skirrow MB, Blaser MJ. Nachamkin I, Blaser MJ, editors. Clinical aspects of Campylobacter infection. Campylobacter. 2000. 2nd ed. Washington: ASM Press;69–88.
19. Spence RP, Bruce IR, McFadden AMJ, Hill FI, Tisdall D, Humphrey S, van der Graaf L, van Bergen MAP, Wagenaar JA. Cross-reaction of a Campylobacter fetus subspecies venerealis real-time PCR. Vet Rec. 2011. 168:131.

20. Thibier M, Guerin B. Hygienic aspects of storage and use of semen for artificial insemination. Anim Reprod Sci. 2000. 62:233–251.

21. Thompson SA, Blaser MJ. Nachamkin I, Blaser MJ, editors. Pathogenesis of Campylobacter fetus infections. Campylobacter. 2000. 2nd ed. Washington: ASM Press;321–347.
22. Tramuta C, Lacerenza D, Zoppi S, Goria M, Dondo A, Ferroglio E, Nebbia P, Rosati S. Development of a set of multiplex standard polymerase chain reaction assays for the identification of infectious agents from aborted bovine clinical samples. J Vet Diagn Invest. 2011. 23:657–664.

23. van Bergen MAP, Simons G, van der Graaf-van Bloois L, van Putten JPM, Rombout J, Wesley I, Wagenaar JA. Amplified fragment length polymorphism based identification of genetic markers and novel PCR assay for differentiation of Campylobacter fetus subspecies. J Med Microbiol. 2005. 54:1217–1224.

24. Véron M, Chatelain R. Taxonomic study of the genus Campylobacter Sebald and Véron and designation of the neotype strain for the type species, Campylobacter fetus (Smith and Taylor) Sebald and Véron. Int J Syst Bacteriol. 1973. 23:122–134.
25. Wang G, Clark CG, Taylor TM, Pucknell C, Barton C, Price L, Woodward DL, Rodgers FG. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J Clin Microbiol. 2002. 40:4744–4747.
26. Willoughby K, Nettleton PF, Quirie M, Maley MA, Foster G, Toszeghy M, Newell DG. A multiplex polymerase chain reaction to detect and differentiate Campylobacter fetus subspecies fetus and Campylobacter fetus-species venerealis: use on UK isolates of C. fetus and other Campylobacter spp. J Appl Microbiol. 2005. 99:758–766.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download