Abstract
T-cell receptor γ alternate reading frame protein (TARP) is expressed by human prostate epithelial, prostate cancer, and mammary cancer cells, but is not found in normal mammary tissue. To date, this protein has only been described in humans. Additionally, no animal model has been established to investigate the potential merits of TARP as tumor marker or a target for adoptive tumor immunotherapy. In this study conducted to characterize feline T-cell receptor γ sequences, constructs very similar to human TARP transcripts were obtained by RACE from the spleen and prostate gland of cats. Transcription of TARP in normal, hyperplastic, and neoplastic feline mammary tissues was evaluated by conventional RT-PCR. In felines similarly to the situation reported in humans, a C-region encoding two open reading frames is spliced to a J-region gene. In contrast to humans, the feline J-region gene was found to be a pseudogene containing a deletion within its recombination signal sequence. Our findings demonstrated that the feline TARP ortholog is transcribed in the prostate gland and mammary tumors but not normal mammary tissues as is the case with human TARP.
The T-cell receptor γ alternate reading frame protein (TARP) is transcribed and expressed by epithelial cells of the prostate gland as well as benign prostate gland tumors, prostate cancer cells, and breast cancer cells in humans [8,9,30]. The transcript is derived from the T-cell receptor γ (TRG) J-region gene, TRGJP in germline configuration, and the constant region gene, TRGC1, of the human TRG locus. This region contains two open reading frames (ORFs) [8] but only the first is translated. The product is a 7-kDa protein with five leucine residues in heptad repeats followed by a basic region forming a potential leucine zipper motif [30]. The protein is located in the outer mitochondrial membrane, and its expression is increased in prostate and breast cancer cells [9,14,30]. Its expression in cancer cells in vitro leads to increased cell growth [29].
Because of TARP expression in prostate and breast cancer, this protein has been tested as a target for cytotoxic T-cells [1,7,19] and helper T-cells [12] as an innovative approach for specific adoptive tumor immunotherapy. Similar truncated transcripts originating from T-cell receptor α genes have been found in human and murine sarcomas. These proteins also seem to be involved in regulating tumor cell proliferation [6]. The TARP promoter has also been used to study adoptive tumor therapy [2-4].
TARP is transcribed from a J-region sequence and C-region sequence in germline configuration. In contrast, normal T-cell receptors are transcribed and translated from somatically rearranged genes of the same locus. This rearrangement of antigen receptor genes is known as V(D)J recombination [18]. Germline transcription of V-region and J-C-region genes takes place during V(D)J recombination within the thymus and their transcripts participate in the regulation of this process [13].
V(D)J recombination is initiated by products of the recombination activating genes 1 and 2 that bind to the recombination signal sequence (RSS) and induce DNA double-strand breaks [15,24]. The signal ends and coding ends then undergo non-homologous end joining [10]. The RSSs are composed of a highly conserved heptamere, a conserved adenine rich nonamere, and a less conserved spacer region of 12 ± 1 or 23 ± 1 bp. The length of the spacer is used to characterize the RSSs, which are annotated as 12RSS and 23RSS, respectively [10,22,23].
In the present study, we identified a feline transcript similar to human TARP composed of a J-region sequence and C-region sequence. This transcript was originally obtained from a splenic cDNA library established to characterize feline TRG sequences. Additionally, transcription of this construct was found in feline mammary neoplasm, similar to humans.
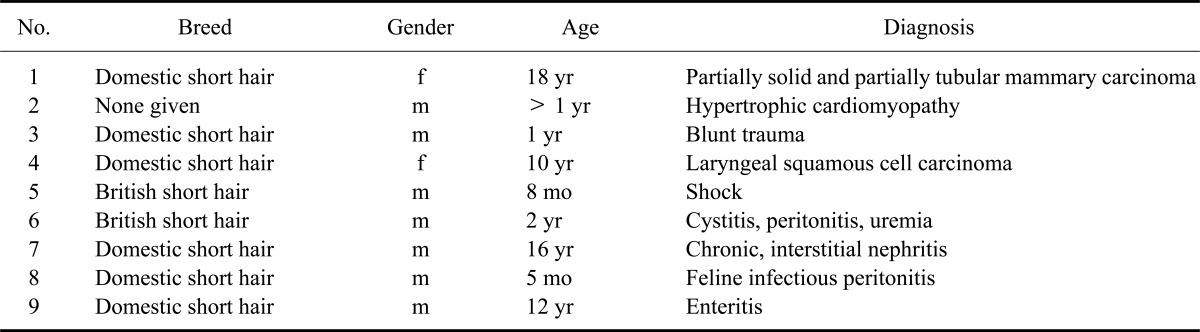
Total RNA was extracted from the spleens of three domestic shorthair cats of different ages and genders (see Table 1, cats Nos. 1~3) and the prostate gland of five male cats (cats Nos. 5~9) submitted for routine pathological evaluation to our departments. Prior to performing rapid amplification of cDNA ends (RACE), total RNA was extracted with a Purescript RNA Isolation Kit (Biozym Scientific, Germany). Genomic DNA was isolated from the same tissues using a Puregene DNA Isolation Kit (Biozym Scientific, Germany). For RT-PCR amplification of presumed feline TARP sequences from feline prostate glands, total RNA was extracted with a High Pure RNA Tissue Kit (F. Hoffmann-La Roche, Germany).
To assess TARP transcription in feline mammary tumors, 32 FFPE neoplastic and 14 normal or hyperplastic mammary tissue samples from 22 female animals submitted for routine diagnostics procedures in 2008 and 2009 were analyzed. Samples of all major tissues from three additional animals excluding the spleen were pooled to evaluate transcription in other tissues. Total RNA was extracted from 30 to 40 paraffin sections of 2 µm thickness each. This was approximately equal to 10 mg of deparaffinized tissue. Tissue sections were deparaffinized by incubating with 800 µL xylene for 5 min at room temperature and pelleting at 14,000 × g at room temperature for 5 min. The resulting pellets were resuspended 300 µL of 99% ethanol and washed twice with 300 µL of 99% ethanol. Proteins were digested by resuspending the pellets in 300 µL cell lysis solution (Gentra Puregene Tissue Kit; Qiagen, Germany) and homogenizing by pipetting up and down. Next, 1.5 µL proteinase K solution (Gentra Puregene Tissue Kit; Qiagen, Germany) was added and the solution was incubated at 55℃ for 1 to 3 h. The samples were then inactivated by incubating at 94℃ for 10 min.
After cooling to room temperature, 800 µL TRIzol Reagent (Invitrogen, USA) were added and the samples were incubated at room temperature for 5 min. Next, 200 µL of chloroform (Carl Roth, Germany) were added. The samples were then incubated at room temperature for 5 min and centrifuged at 14,000 × g for 5 min at 4℃.
To remove residual phenol from the samples, an additional 400 µL of chloroform were added to the upper phase. After phase separation, 600 µL of isopropanol (Carl Roth, Germany) were added to the upper phase and samples were pelleted at 14,000 × g and 4℃ for 5 min. The pellets washed twice with 500 µL of 75% ethanol (Carl Roth, Germany), air dried, and resuspended in 20 µL diethylpyrocarbonate-treated water (Carl Roth, Germany).
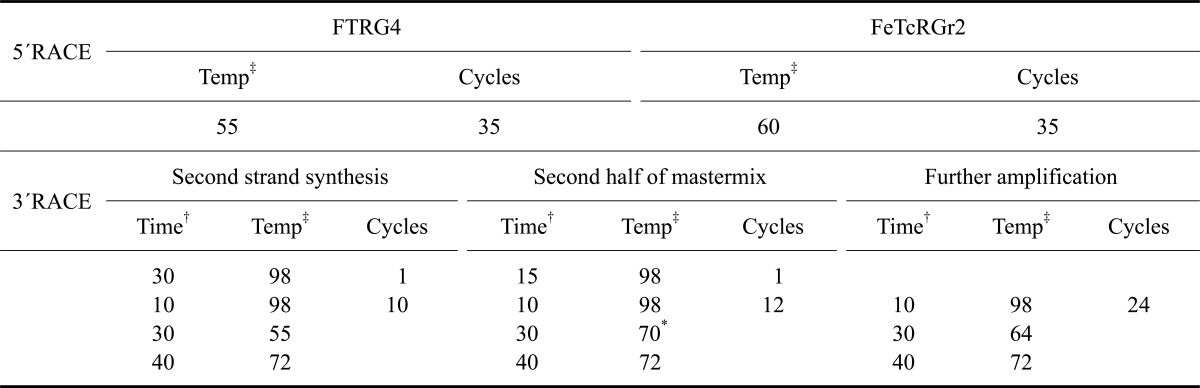
5'RACE was performed using a 5'/3' RACE Kit (F. Hoffmann-La Roche, Germany) and a Multicycler PTC 200 Gradient (Biozym, Germany). Amplification was carried out by nested PCR using Phusion High-Fidelity DNA Polymerase (BioCat, Germany) to ensure high sequence accuracy. The total reaction volume was 50 µL containing 1.5 mM MgCl2, 0.2 mM of each dNTP (Invitrogen, USA), 0.2 µM of each primer, 0.02 U/µL polymerase and 1 µg RNA. Primers were designed according to previously published sequences (D89023) [5]. FeTcRGr3 was used as a primer for cDNA synthesis, and FTGR4 along with FeTcRGr2 were used for amplification (for primer sequences, see Table 2). Primer annealing locations and the location of cloned sequences are shown in Fig. 1. DNA was denatured for 30 sec at 98℃, each cycle consisted of melting for 10 sec at 98℃, annealing for 30 sec, and elongation for 30 sec at 72℃ (for reaction conditions see Table 3). This was followed by a final elongation for 5 min at 72℃. PCR products were cloned with the AccepTor Vector Kit (Merck, Germany) and sequenced by a commercial laboratory (Eurofins MWG Operon, Germany) as previously described [26].
3'RACE was performed using a 5'/3' RACE Kit (F. Hoffmann-La Roche, Germany) a Multicycler PTC 200 Gradient (Biozym, Germany). Amplification was catalyzed by Phusion High-Fidelity DNA Polymerase (BioCat, Germany) and performed in two steps. First, second-strand synthesis was conducted using the first half of a 50-µL PCR master mix composed as listed above containing 0.1 µM fTARPf1 as the gene-specific primer. After ten PCR cycles, the other half of the aforementioned master mix was added along with 0.24 µM of the second gene-specific primer fTARPf2 and 0.16 µM of the RACE anchor primer. Reaction conditions are listed in Table 3. The PCR products were cloned and sequenced as described above.
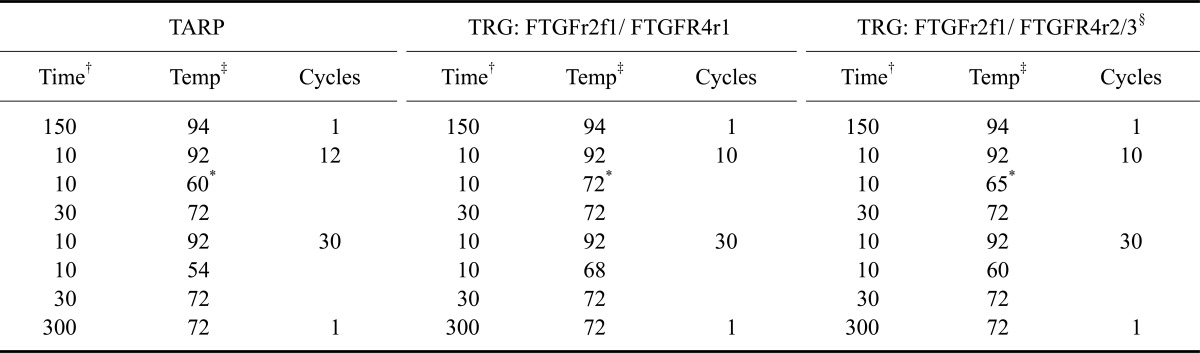
cDNA was synthesized using a RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA) using the included random hexamer primers as recommended by the manufacturer's instructions. GoTaq DNA Polymerase (Promega, USA) was used to amplify cDNA and genomic DNA. The PCR reaction contained 100 ng DNA, 2.0 mM MgCl2, 0.2 mM of each dNTP, 0.2 µM of each primer, and 0.025 U/µL DNA polymerase. The final reaction volume was 25 µL and the PCR conditions are listed in Table 4. fTARPf1 and -2 were used as forward primers; FTGJr1, fTARPr1, fTARPr2, and FTGPIr1 were used as reverse primers.
Sequences were analyzed using the Basic Local Alignment Search Tool (National Center for Biotechnology Information, USA) and ClustalW (UCD Dublin, Ireland). GeneDoc 2.6.003 (Pittsburgh Supercomputing Center, USA) was used for displaying multiple sequence alignments. The sequences generated have been submitted to the Nucleotide Sequence Database (European Molecular Biology Laboratory, Germany): AM502837~AM502846, AM940997 (complete CDS), and FN433011.
Approximately 500 ng of total RNA from each sample were reverse transcribed using RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA). A PCR master mix of the same composition as described above was used. The fTARPf2 and fTARPr3 primers were used and the reaction conditions were slightly modified. Starting temperature for annealing was 63℃ followed by a touch down of 1℃ per cycle for 10 cylcles. Afterwards 57℃ were used. cDNA amplification was monitored using the YWHAZ housekeeping gene as previously described [20].
The seven clones (AM502839~AM502845) produced by a classical 5'RACE procedure using total RNA extracted from the spleens of two different cats (Nos. 2 and 3) included a transcript similar to human TARP (Fig. 2A). This transcript consisted of a TRG J-region sequence that was spliced to a C-region sequence (Fig. 1). To analyze the whole sequence of the transcript, 3'RACE was conducted with primers specific for the J-region sequence (cat No. 1). Resulting clones could only be sequenced in the forward direction. Sequencing with the reverse primer came to a halt within the poly-A-tail. Therefore, approximately 70 bp at the 3' end have not been sequenced. However, the sequence was almost identical (over 99% homology) to fTRGC4 that has been sequenced previously (GenBank accession No. AM489512). fTARP was also cloned and sequenced from total splenic RNA of cat No. 1 using fTARPf1 and FeTcRGr3/fTARPr2 (primers Nos. 4 and 2 shown in Fig. 1). This amplicon had more than 99% identity to the clones from animals Nos. 2 and 3.
To measure TARP transcription in the feline prostate gland, fTARP was amplified and sequenced from total prostatic RNA of five animals (Nos. 5 through 9; Fig. 3C) using the same primers. These amplicons also had more than a 99% identity to the sequenced clones. The C-region sequence encoded two ORFs. The first one started at the beginning of the C-region, contained 189 bp, and was out of frame compared to the fTRG transcripts. The second started at an 'ATG' site 185 bp downstream from the beginning of the C-region and contained 462 bp. This second ORF was in frame compared to the fTRG transcripts.
Sequence similarity of the coding sequence for human and feline TARP was 84% at the nucleotide level and 70% at the amino acid level (Fig. 2A). Sequence similarity of the second ORF between cats and humans was approximately 30% at the nucleotide level. The included J-region sequences had a 37% sequence similarity.
All feline C-region genes except for TARP/TRGC4 contained a start codon at the same position as human TARP. In TARP/TRGC4, a 'TTG' codon was found in this position. However, there was a start codon precisely at the beginning of the C-region. This position was located 12 bp upstream of the 'TTG' codon.
Similarity of the J-region sequence was 76% to fTRGJ2.1, 83% to fTRGJ2.2, 88% to fTRGJ2.3, and 80% to fTRGJ2.4. The J-region sequence in fTARP had a 19-bp deletion at the position of the 12RSS (Fig. 2B). This J-region was therefore named fTRGJP. The sequence, including a 5' intron, was also found in feline genomic sequences (gnl|ti|755161287) from the Trace Archive Nucleotide BLAST database (NCBI, USA).
To determine the genomic organization of feline TARP, the fTARPf1, FTGJr1, and fTARPr1 primers were designed. An amplicon recovered from genomic DNA using fTARPf1 and FTGJr1 was identified in all animals tested (Fig. 3A). The amplicon produced by fTARPr1, indicating the presence of spliced TARP DNA within the genome, was produced from the DNA of only one animal (No. 4; Fig. 3B). Additionally, the FTGPIr1 primer was specific for the intron downstream of fTRGJP. The amplicon produced by this primer was also recovered in all animals tested (Fig. 3A).
For cat No. 4, we amplified DNA pretreated with DNase and RNase using fTARPf1 and fTARPr1 or FTGPIr1. No product was generated from the DNase-treated DNA, indicating that both variants are present in genomic DNA (Fig. 3B). The amplicons generated by fTARPf1 and fTARPr1 or FTGPIr1 were cloned and sequenced. Similarity to fTARP was approximately 99% for both the genomic and cDNA sequences. Weak transcription of recombined TRG-V domains was detected within the prostate of cats Nos. 5~9 with the primers fTRGFR2f1 and fTRGFR4r1 (data not shown). Additional primer combinations yielded no amplicons.
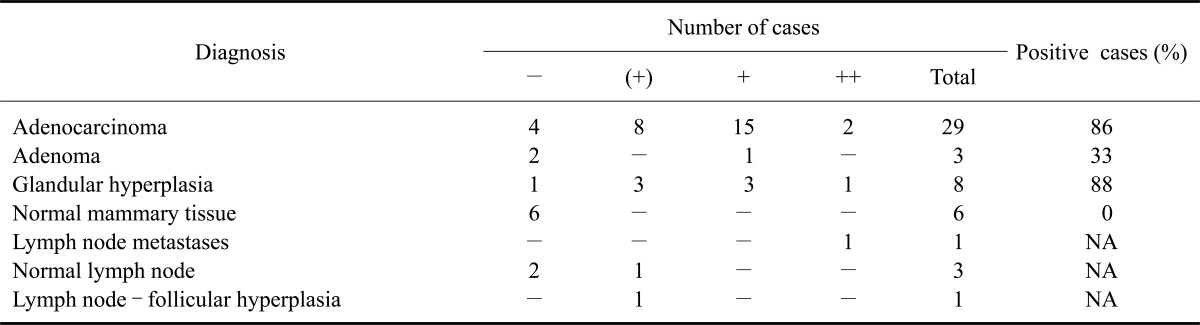
The amount of total RNA extracted from FFPE mammary tissues ranged from 0.6 to 46 µg (average: 8.6 µg, median: 5 µg, average 260/280 ratio: 1.6). Results of RT-PCR amplification of feline TARP sequences are summarized in Table 5. Because band density varied considerably, the results are presented in a semi-quantitative manner (Fig. 3D). The majority (81%) of the neoplastic tissues expressed TARP sequences whereas this expression was not observed in any of the normal mammary gland samples. No TARP mRNA was detected in the pooled RNA samples from other tissues of the three cats tested. All samples yielded bands for YWHAZ of approximately equal density.
Transcription of a J-region sequence spliced to a C-region sequence is typical for human TARP. We detected the same constellation for one feline C-region and J-region sequence, suggesting that this transcript is the feline counterpart of human TARP. Whether these genes are located within the feline TRG locus, as is the case of human TARP [8,30], or comprise a separate locus that arose by gene duplication is not certain at the moment. Interestingly, the C-region sequence involved (fTRGC4) was never detected in combination with a functionally recombined variable domain [26-28]. Thus, we could not demonstrate its utilization as an fTRGC region. Furthermore, this C-region was associated with a carnivore-specific short interspersed element (CAN-SINE) located 468 bp in the 3' direction [27].
Due to its homology with rabbit TRG cDNA sequences (D38136, 86% nucleic acid identity), we first speculated that the presumed TARP we amplified could be a recombined VJ sequence. However, the sequence specific for the fTARPf1 primer is located within the intron upstream of TRGJP. An amplicon can thus be produced from genomic DNA using this primer and FTGJr1.
Interestingly, the start codon in this sequence was found 12 bp upstream from the corresponding position in human TARP. This indicates that after mutation of the original start codon present in all other feline C-region genes, a second mutation must have occurred. Therefore, fTRGC4 might have another role than that specific for a C-region of TRG.
The associated J-region sequence in fTARP features a 19-bp deletion within its RSS. fTRGJP is thus a pseudogene with regard to the TRG locus since productive recombination with a V-region is prevented. TR and immunoglobulin loci of all species tested so far contain multiple pseudogenes [10,13]. Pseudogenes arise from point mutations, deletions, or insertions after the regions have been duplicated. A pseudogene results if these mutations either alter the RSS or cause a reading frame shift [17,21].
Another possible manner in which the presumed fTARP transcript was formed, at least within splenic tissue, could be transcription of this sequence in a germline configuration. Each gene that encodes a region of the antigen receptors has its own promoter that is probably necessary for successful recombination [11]. These promoters induce transcription prior to recombination [10]. V-region and J-C-region genes are transcribed in germline configuration during recombination, but this takes place within the thymus in pre-T-cells and is induced by IL-7 signaling [13]. It is therefore unlikely that the alleged TARP transcript in spleen and prostate gland is caused by germline transcription. However, transcription of J-region genes in germline configuration has been observed in the spleen of fetal and adult cattle, but it has not been determined whether these transcripts could resemble TARP [16]. Excluding fTRGJP, we did not detect any transcription of fTRG sequences in germline configuration in feline spleen tissue. With regard to fTARP, this J-region sequence is only part of the 5' untranslated region.
Additionally, fTARP was amplified in cat No. 4 from genomic DNA in "spliced form". This finding indicates that in some animals fTARP additionally exists as a processed and reintegrated pseudogene. The fact that we found this pseudo gene in only one of the animals studied indicates that this transcript was very recently reintegrated into the cat's genome.
Similar to humans, alleged feline TARP transcripts were present in feline mammary adenocarcinomas and adenomas in the current study. However, cases of glandular hyperplasia were also positive for TARP transcription. This has not been reported in humans. Further testing should be conducted to determine whether TARP transcription indicates potential progression towards neoplastic transformation. Additionally, weak transcription found in lymphatic tissues should be further investigated because this might hamper the use of fTARP in tumor immunotherapy.
In summary, we have described an fTRC region highly transcribed in combination with an fTRGJ pseudogene in feline spleen and prostate as well as neoplastic tissues from feline mammary glands. The molecular characteristics were highly similar to those of human TARP. Further characterization of this transcript in cats could lead to a better understanding of its physiological function and significance. Additionally, if this mRNA sequence is also translated in feline cancer cells, it could serve as a tumor marker or a target for anti-cancer immunotherapy in this species. Furthermore, characterization of TARP in cats may result in a suitable animal model to analyze its potential as target for adoptive tumor immunotherapy.
Acknowledgments
This work was supported by a "Graduiertenstipendium" of the Justus-Liebig-University Giessen and by the PhD Programme of the Faculties of Veterinary Medicine and Medicine of Justus Liebig University, Giessen, Germany. The authors thank Josephine Braun (Department of Veterinary Pathology, Freie Universität Berlin, Berlin, Germany) for editing this manuscript.
References
1. Carlsson B, Tötterman TH, Essand M. Generation of cytotoxic T lymphocytes specific for the prostate and breast tissue antigen TARP. Prostate. 2004; 61:161–170. PMID: 15305339.

2. Cheng WS, Dzojic H, Nilsson B, Tötterman TH, Essand M. An oncolytic conditionally replicating adenovirus for hormone-dependent and hormone-independent prostate cancer. Cancer Gene Ther. 2006; 13:13–20. PMID: 16052227.

3. Cheng WS, Giandomenico V, Pastan I, Essand M. Characterization of the androgen-regulated prostate-specific T cell receptor γ-chain alternate reading frame protein (TARP) promoter. Endocrinology. 2003; 144:3433–3440. PMID: 12865322.

4. Cheng WS, Kraaij R, Nilsson B, van der Weel L, de Ridder CMA, Tötterman TH, Essand M. A novel TARP-promoter-based adenovirus against hormone-dependent and hormone-refractory prostate cancer. Mol Ther. 2004; 10:355–364. PMID: 15294182.

5. Cho K, Youn HY, Okuda M, Satoh H, Cevario S, O'Brien SJ, Watari T, Tsujimoto H, Hasegawa A. Cloning and mapping of cat (Felis catus) immunoglobulin and T-cell receptor genes. Immunogenetics. 1998; 47:226–233. PMID: 9435341.
6. Croci S, Strippoli P, Bonsi L, Bagnara GP, Guizzunti G, Sartini R, Tonelli R, Messina C, Pierdomenico L, Lollini PL. Expression of T cell receptor alpha gene (TCRA) in human rhabdomyosarcoma and other musculo-skeletal sarcomas. Gene. 2005; 353:16–22. PMID: 15935573.

7. Epel M, Carmi I, Soueid-Baumgarten S, Oh SK, Bera T, Pastan I, Berzofsky J, Reiter Y. Targeting TARP, a novel breast and prostate tumor-associated antigen, with T cell receptor-like human recombinant antibodies. Eur J Immunol. 2008; 38:1706–1720. PMID: 18446790.

8. Essand M, Vasmatzis G, Brinkmann U, Duray P, Lee B, Pastan I. High expression of a specific T-cell receptor γ transcript in epithelial cells of the prostate. Proc Natl Acad Sci USA. 1999; 96:9287–9292. PMID: 10430935.

9. Fritzsche FR, Stephan C, Gerhardt J, Lein M, Hofmann I, Jung K, Dietel M, Kristiansen G. Diagnostic and prognostic value of T-cell receptor gamma alternative reading frame protein (TARP) expression in prostate cancer. Histol Histopathol. 2010; 25:733–739. PMID: 20376779.
10. Jung D, Alt FW. Unraveling V(D)J recombination: insights into gene regulation. Cell. 2004; 116:299–311. PMID: 14744439.
11. Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006; 24:541–570. PMID: 16551259.

12. Kobayashi H, Nagato T, Oikawa K, Sato K, Kimura S, Aoki N, Omiya R, Tateno M, Celis E. Recognition of prostate and breast tumor cells by helper T lymphocytes specific for a prostate and breast tumor-associated antigen, TARP. Clin Cancer Res. 2005; 11:3869–3878. PMID: 15897588.

13. Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol. 2009; 21:133–139. PMID: 19362456.

14. Maeda H, Nagata S, Wolfgang CD, Bratthauer GL, Bera TK, Pastan I. The T cell receptor γ chain alternate reading frame protein (TARP), a prostate-specific protein localized in mitochondria. J Biol Chem. 2004; 279:24561–24568. PMID: 15150260.

15. McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995; 83:387–395. PMID: 8521468.

16. Miccoli MC, Vaccarelli G, Lanave C, Cribiu EP, Ciccarese S. Comparative analyses of sheep and human TRG joining regions: evolution of J genes in Bovidae is driven by sequence conservation in their promoters for germline transcription. Gene. 2005; 355:67–78. PMID: 16039073.
17. Mighell AJ, Smith NR, Robinson PA, Markham AF. Vertebrate pseudogenes. FEBS Lett. 2000; 468:109–114. PMID: 10692568.

19. Oh S, Terabe M, Pendleton CD, Bhattacharyya A, Bera TK, Epel M, Reiter Y, Phillips J, Linehan WM, Kasten-Sportes C, Pastan I, Berzofsky JA. Human CTLs to wild-type and enhanced epitopes of a novel prostate and breast tumor-associated protein, TARP, lyse human breast cancer cells. Cancer Res. 2004; 64:2610–2618. PMID: 15059918.

20. Penning LC, Vrieling HE, Brinkhof B, Riemers FM, Rothuizen J, Rutteman GR, Hazewinkel HAW. A validation of 10 feline reference genes for gene expression measurements in snap-frozen tissues. Vet Immunol Immunopathol. 2007; 120:212–222. PMID: 17904230.

21. Rothenfluh HS, Blanden RV, Steele EJ. Evolution of V genes: DNA sequence structure of functional germline genes and pseudogenes. Immunogenetics. 1995; 42:159–171. PMID: 7642227.

22. Sakano H, Hüppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979; 280:288–294. PMID: 111144.

24. van Gent DC, McBlane JF, Ramsden DA, Sadofsky MJ, Hesse JE, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995; 81:925–934. PMID: 7781069.

25. Weiss ATA, Delcour NM, Meyer A, Klopfleisch R. Efficient and cost-effective extraction of genomic DNA from formalin-fixed and paraffin-embedded tissues. Vet Pathol. 2011; 48:834–838. PMID: 20817894.

26. Weiss ATA, Hecht W, Henrich M, Reinacher M. Characterization of C-, J- and V-region-genes of the feline T-cell receptor γ. Vet Immunol Immunopathol. 2008; 124:63–74. PMID: 18456341.

27. Weiss ATA, Hecht W, Reinacher M. Feline T-cell receptor γ V- and J-region sequences retrieved from the trace archive and from transcriptome analysis of cats. Vet Med Int. 2010; 2010:953272. PMID: 20634910.
28. Weiss ATA, Klopfleisch R, Gruber AD. T-cell receptor γ chain variable and joining region genes of subgroup 1 are clonally rearranged in feline B- and T-cell lymphoma. J Comp Pathol. 2011; 144:123–134. PMID: 20846665.

29. Wolfgang CD, Essand M, Lee B, Pastan I. T-cell receptor γ chain alternate reading frame protein (TARP) expression in prostate cancer cells leads to an increased growth rate and induction of caveolins and amphiregulin. Cancer Res. 2001; 61:8122–8126. PMID: 11719440.
30. Wolfgang CD, Essand M, Vincent JJ, Lee B, Pastan I. TARP: a nuclear protein expressed in prostate and breast cancer cells derived from an alternate reading frame of the T cell receptor γ chain locus. Proc Natl Acad Sci USA. 2000; 97:9437–9442. PMID: 10931945.
Fig. 1
Portion of the feline T-cell receptor γ (fTRG) locus containing the hypothetical feline T-cell receptor γ alternate reading frame protein (fTARP) and TARP mRNA (not to scale). Alternatively, two loci may exist: one containing fTRGC4 and the carnivore-specific short interspersed nuclear elements (CAN-SINE), and a second one containing fTARP and fTRGJP. Arrows and numbers indicate primer position and orientation. 1: FeTcRGr2, 2: FeTcRGr3/fTARPr2, 3: FTGR4, 4: FTGVf1/fTARPf1, 5: fTARPf2, 6: FTGJr1, 7: FTGCr1/fTARPr1, fTARPr3, 8: FTGPIr1, C.-S.: CAN-SINE, 5'UTR: 5'untranslated region, 3'UTR: 3'untranslated region, AAAAA: poly-A-tail.
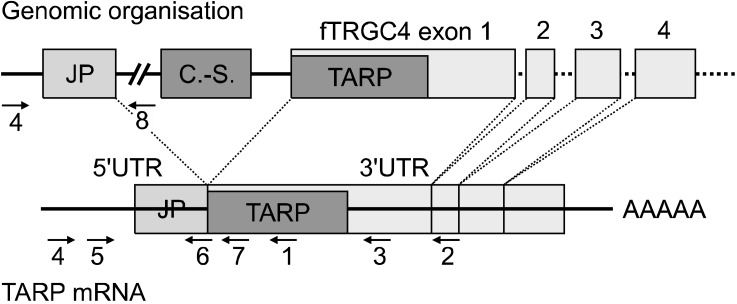
Fig. 2
(A) Alignment of the deduced amino acid sequences of human and putative feline TARP. In human TARP (hTARP), five leucine residues in heptad repeats can be found. In fTARP, only four leucine residues were observed. The basic region downstream of the last leucine displayed high homology. (B) Alignment of clone pFTGII.164, the genomic sequence of fTRGJP (gnl|ti|755161287), and fTRGJ2.3 (gnl|ti|915242736). In fTRGJP, the heptamer of the 12RSS was deleted. The spacer of the fTRGJ2.3 12RSS was either 10- or 11-bp long, or the nonamer ended on 'T'.
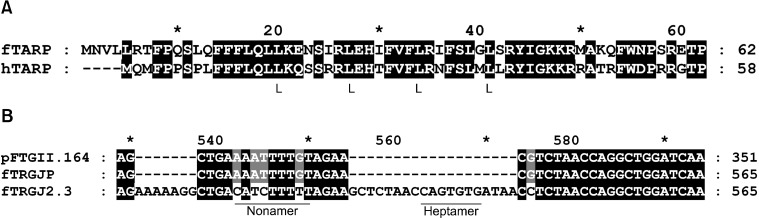
Fig. 3
PCR amplification of feline TARP with different primer sets from genomic DNA and cDNA. (A) TARP amplification of genomic DNA from five different cats using FTGVf1/fTARPf1 as the forward primer and FTGJr1 (lanes 1, 11, 21, 31, and 41 are specific for fTRGJP), FTGCr1/fTARPr1 (lanes 2, 12, 22, 32, and 42 are specific for spliced TARP) and FTGPIr1 (lanes 3, 13, 23, 33, and 43 specific for the intron downstream of fTRGJP) as the reverse primers. Lane 0: control (no template), lane M: DNA size marker. (B) Amplification of DNA from cat No. 4 using FTGVf1/fTARPf1 and FTGCr1/fTARPr1 (lanes 1~4 are specific for spliced TARP) or FTGPIr1 (lanes 11~14 are specific for the intron downstream of fTRGJP). DNA was untreated (lanes 1 and 11), treated with RNase (lanes 2, 3, 12 and 13), and treated with DNase (lanes 4 and 14). Lane 0: control (no template), lane M: DNA size marker. (C) Amplification of TARP cDNA/mRNA from feline prostate glands (primers: fTARPf1 and fTARPr2). Lane 1: cat No. 5, lane 2: cat No. 6, lane 3: cat No. 7, lane 4: cat No. 8, lane 5: cat No. 9, lane 0: control (no template). (D) Amplification of TARP cDNA/mRNA with fTARPf2 and fTARPr3 primers from normal mammary gland (lanes 1, 2, and 5), mammary glandular hyperplasia (lane 3), mammary adenomas (lanes 14 and 15), mammary adenocarcinomas (lanes 4, 6, 7, 9, 10, 11, and 12), normal lymph node (lane 13), and lymph node metastases of mammary adenocarcinoma (lane 8). Lane M: DNA size marker, lane 0: control (no template), lane +: positive control.
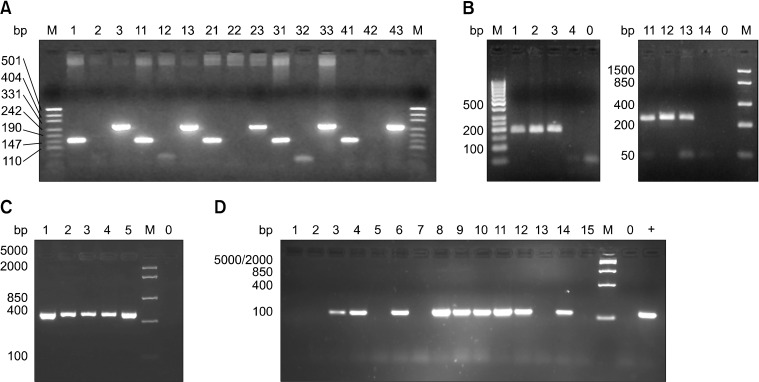
Table 2
Primer sequences and positions
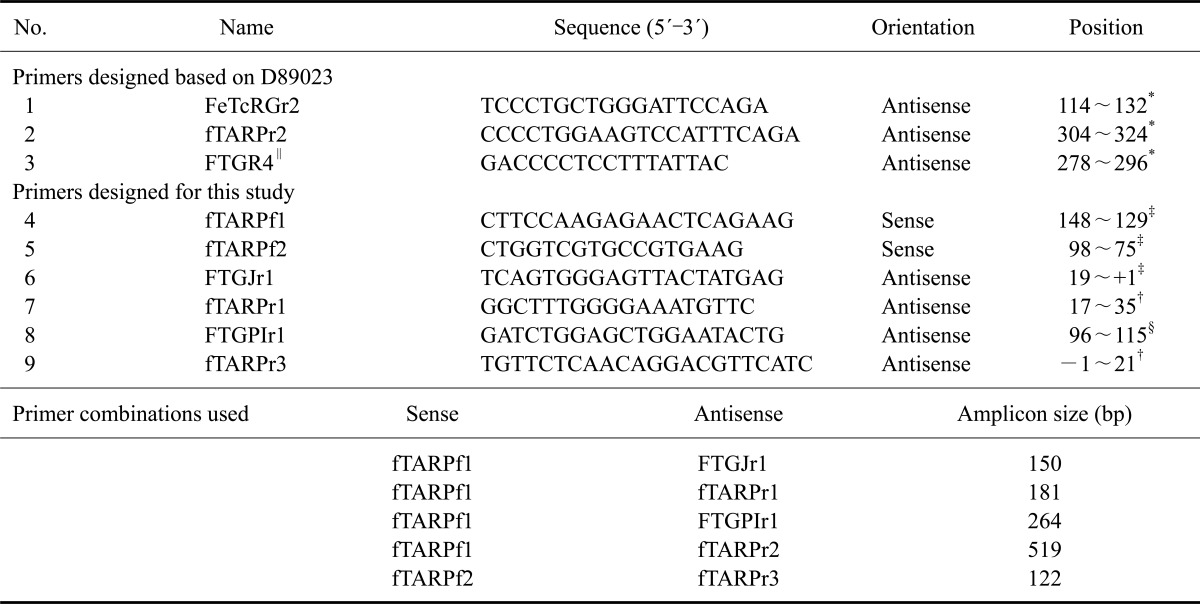
*Position within D89023. †Position within feline T-cell receptor γ constant region gene (fTRGC). ‡bp upstream of the C-region in the spliced form (cDNA). §bp downstream of feline TRG J-region gene (fTRGJP). ∥presented in Cho et al. [5].




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download