Abstract
The present study describes the genotypic distribution of rotaviruses (RVs) in an Indian bovine population with unexpectedly higher proportions of G3 alone or in combination of G8/G10. PCR-genotyping confirmed that 39.4% (13/33) of the prevalent RVs were the G3 type while 60.6% (20/33) were dual G3G10 or G3G8 types. P typing revealed that 93.9% (31/33) of the samples were P[11] while 6.1% (2/33) possessed a dual P[1]P[11] type. Sequence analysis of the VP7 gene from G3 strains viz. B-46, 0970, and BR-133 showed that these strains had sequence identities of 90.5% to 100% with other bovine G3 strains. The highest identity (98.9% to 100%) was observed with RUBV3 bovine G3 strains from eastern India. The G3 strains (B-46, 0970, and BR-133) showed 97.5% to 98.8% sequence homologies with the Indian equine RV strain Erv-80. Phylogenetic analysis demonstrated that G3 strains clustered with bovine RUBV3 and J-63, and equine Erv-80 G3. Overall, these results confirmed that the incidence of infection by RVs with the G3 genotype and mixed genotypes in the bovine population was higher than previously predicted. This finding reinforces the importance of constantly monitoring circulating viral strains with the G3 genotype in future surveillance studies.
Rotavirus (RV) is one of the leading causes of diarrhea in neonatal animals, humans, and birds. Infection with RVs has resulted in colossal economic losses in the livestock industry [8]. These losses are much greater in developing countries like India where socio-economic conditions favor the survival of virus in the environment [11]. RVs are icosahedral non-enveloped viruses belonging to the family Reoviridae with a genome comprising of 11 segments of double-stranded (ds) RNA, each of which encodes one of the six structural (VP1~4, 6, and 7) or six non-structural (NSP1-6) proteins [7]. The complete infectious RV virion consists of three concentric layers: the inner core, inner capsid, and outer capsid [7]. Three out of the six structural proteins (VP4, VP6, and VP7) have major antigenic determinants and are used for RV classification into groups (A~G), sub-groups (SGI/II), and genotypes/serotypes (G&P) [7]. Group A rotavirus (RVA) is the most predominant serogroup present in bovines and other mammalian species throughout the world [7].
Based on neutralizing antibodies against the outer capsid proteins VP7 and VP4, two independent glycoprotein (G) and protease sensitivity (P) serotype systems have been established. However, sequence-based genotyping [4,11,12,14,15,30] targeting the VP7 (G type) and VP4 (P type) protein genes has become more a common method for RVA typing due to difficulties in generating serotyping reagents and performing the time-consuming assays. To date, 25 G (G1~25) types and 32 P (P[1~32]) types have been identified for different RV strains [3,6,19,32]. Genotypes in RVs circulating among different animal populations have been examined in order to design effective vaccines and assess RV cross-species infectivity. Changing patterns in prevalence of bovine G and P types have been observed over time. The most predominant genotypes in bovines are G6, G8, and G10 with sporadic incidences of G1, G3, G5, or G15. The most prevalent P types are P[1], P[5], and P[11] with sporadic occurrence of P[14], P[15], P[17], and P[21] [2,7,9,11,17,21,23,25,26].
In India, G10 (50~60%) has been the predominant bovine RV genotype over the last 10 years followed by G6 (25~35%) [11,14,20,22,27,28,34]. Varshney et al. [33] were the first to report on the existence of G3 RV in buffalo calves (10.2%) in central India. This was the second most predominant strain after G10. In 2007, Ghosh et al. [11] also reported a 3% incidence of G3 RVs in claves from eastern India, indicating that either the number of bovines infected with RV G3 genotypes are increasing or these animals have not been taken into account while conducting genotyping surveys in the country [14,20,22,27,28,34]. The present study describes G and P genotypes prevalent in Indian bovine populations from diverse tropical, sub-tropical/semi-arid, and temperate western Himalayan regions during 2007~2010. Sequence and phylogenetic analyses of the G3 genotype presently circulating in the country were also performed.
A total of 378 fecal samples were collected from cattle and buffalo calves up to 3 months old that were suffering from diarrhea. These animals belonged to organized and unorganized dairy farms in different ecoregions of India including temperate western Himalaya (Tarai and foothills of Himalaya, Uttarakhand state), tropical, semi-tropical, and semi-arid regions (Haryana, Uttar Pradesh, and Madhya Pradesh) of India. A 10% fecal suspension (w/v) was prepared with phosphate buffered saline (0.01 M, pH 7.4; Sigma, USA). Samples were centrifuged at 2,000 × g for 10 min to remove coarse particulate matter and the upper aqueous layer was filtered through a 0.22-µm pore filter (MDI, India) into a fresh tube. The suspension was stored at -20℃ until further use. Reference NCDV (G6P[1]) and UK (G6P[5]) cell culture-adapted bovine RVA strains were obtained from the National Research Centre on Equine (India). RNA extracted from these samples was used to optimize reverse-transcription (RT)-PCR and genotyping conditions.
Total RNA was extracted from 500 µL of fecal suspension using an equal volume of TriReagent-LS (Sigma, USA) following the manufacturer's instructions. RNA was dissolved in nuclease-free water (NFW) (Life Technologies, USA) in a final volume of 25 µL. RNA quantity and quality was assessed using a Nanodrop Spectrophotometer (ND-1000; Thermo Scientific, USA) and stored at -20℃ until further use.
The extracted viral RNA was analyzed by PAGE performed as previously described by Laemmli [18] by using 5% stacking and 10% resolving gels. Briefly, extracted RNA (200~500 ng) was mixed with an equal amount of 2× RNA loading buffer (Thermo Fisher Scientific, USA) and loaded into the wells of the gels. Electrophoresis was performed at 120 V in 1× Tris-glycine buffer (25 mM Tris, 192 mM glycine, pH 8.3). The gel was subsequently stained with silver nitrate as previously described by Svensson et al. [29]. All the samples were initially screened with PAGE.
RT of the viral RNA was performed using a random hexamer primer (0.2 µg/µL; Thermo Fisher Scientific, USA). Initially, 50~100 ng of viral RNA, 0.5 µL random hexamer primer, and 2 µL dimethyl sulphoxide (DMSO) were combined in a thin-walled 0.5-mL PCR tube (GenAxy, India). The reaction mixture was incubated at 95℃ for 5 min to denature the dsRNA strands and immediately snap chilled on ice. Next, 4 µL of 5× RT buffer (Promega, USA), 2 µL of 10 mM dNTPs (Thermo Fisher Scientific, USA), 40 U of RNase inhibitor (Life Technologies, USA), and 200 U of Moloney murine leukemia virus reverse transcriptase (Promega, USA) were added and the mixture was incubated at 37℃ in a thermocycler for 1 h (HR-PCR-96G; Haier, China). The reverse transcriptase was denatured at 80℃ for 5 min at the end of the incubation step.
The first-round of PCR amplification of the VP7 gene was performed with 3 µL cDNA, 5 µL 10× PCR buffer, 2 mM MgCl2, 2 µL DMSO, 1 µL dNTPs, 50 pmol of forward and reverse primers (Table 1), and 1.25 U of Taq DNA polymerase (Thermo Fisher Scientific, USA). The total reaction volume was brought to 50 µL with NFW. The first-round of amplification of full-length VP7 (1062 bp) consisted of an initial denaturation at 94℃ for 4 min followed by 35 cycles of 1 min at 94℃, 2 min at 46℃, and 2 min at 72℃, and final incubation at 72℃ for 10 min. A similar PCR protocol was performed to amplify the partial-length VP4 gene (864 bp) for the first-round with the exception that the annealing temperature was set at 48℃. The PCR products were separated by conventional agarose gel electrophoresis (1%, w/v) at 100 V for 1 h in 1× TAE buffer with 0.5 µg/mL ethidium bromide, viewed with a UV transilluminator, and documented using Syngene gel documentation system (Syngene, UK).
Multiplex nested PCR genotype determination was performed as per the method and primers used by Gouvea et al. [12] for G3 (aET3) and G8 (aAT8), Gouvea et al. [13] for G5 and G6, and Iturriza-Gómara et al. [16] for G10. Primers used for the first-round of amplification and subsequent nested PCR for genotyping both the VP4 and VP7 genes are presented in Table 1. Genotyping was performed using 3 µL of the amplicons (1 : 100 dilution) from the first-round of PCR amplification and the typing primers (G3, 5, 6, 8, and 10) with the reverse primer used for generating the first-round VP7 gene product. Amplicons from the first-round of PCR (1 : 100 dilution) and the typing primers (P[1], [5], and [11]) with the forward primer (Bov4Com5) used for generating the first-round VP4 gene product were used for P typing. A diagrammatic sketch with specific amplicon size for the G and P types is presented in Fig. 1. PCR cycling conditions for the second-round were same as those of the first-round. The amplicons were analyzed as described above.
Selected bovine RVA isolates (n = 3) with the G3 genotype from cattle in different geographical areas were amplified as described above. The amplified PCR products were excised from the gel and purified with a High Pure PCR Product Purification kit (Roche Diagnostics, Germany) with final elution in 20 µL of elution buffer. The eluted PCR products were quantified in Nanodrop Spectrophotometer and sequenced with an automated sequencer (ABI 3130, USA) at the Foot and Mouth Disease Typing Facility Lab, Project Directorate on Foot and Mouth Disease, Indian Veterinary Research Institute, Mukteswar (India). Sequence data for these samples were entered into the NCBI GenBank database under the following accession numbers: B-46 (HM235510), BR-133 (JF 720877), and 0970 (HQ 199897).
The sequence chromatogram was visualized with sequencing analysis software (ABI, USA). MegaBLAST (NCBI, USA) was used to analyze the predicted sequence within the non-redundant nucleotide database and confirm the presence of the gene specific for RVs. RotaC web-based RV classification software (Katholieke Universiteit Leuven, Belgium) was also used for sequence confirmation. RVA sequences from animals in different geographical locations within India and the rest of the world were downloaded from the NCBI nucleotide database. Sequence analysis was performed using the LaserGene software package (DNASTAR, USA). Percent identity and divergence of the nucleotide and predicted amino acid sequences were determined to calculate the percentage homology with other G3 RV sequences from different geographical locations. A phylogenetic tree was constructed using Mega 4.0 software [30].
PAGE analyses detected the RV genome in 33 out 378 diarrheic fecal samples from bovine calves that were up to 3 months old. All PAGE-positive RV samples showed a typical 4 : 2 : 3 : 2 "long" electropherogram migration pattern, characteristic of RVA. In the majority of the samples, genome segments 7, 8, and 9 co-migrated as a triplet. PAGE-positive samples were also confirmed by using RT-PCR targeting the group A-specific VP6 gene.
First-round amplification with generic primers of VP7 and VP4 genes produced full-length 1062-bp amplicons for the VP7 gene and partial-length 864-bp products for the VP4 gene in all 33 samples. G typing produced amplicons 373, 581, 884, and 396 bps in size, indicating the presence of G3, G6, G8, and G10 genotypes, respectively (Fig. 2A). None of the samples were found to have the G5 or G6 type RV. P typing of the samples produced amplicons 334, 453, and 661 bp in size, corresponding to P[11], P[1], and P[5] types, respectively (Fig. 2B). However, P[5] was not detected in any of the field isolates (Fig. 2B). A number of samples were confirmed to possess dual G types, either G3G8 or G3G10. The distribution of different combination of genotypes found throughout the study is summarized in Table 2. Typing primers (G&P) also confirmed the reference bovine RVA as G6P[1] for the NCDV strain and G6P[5] for the UK strain (Figs. 2A and B). G3 was found to be the main G type circulating among the ecoregions of India we examined between 2007 ~ 2010 with both single and/or dual infections (G3G8 or G3G10) observed (Fig. 2A). Overall, 13 of the 33 typed samples (39.4%) were identified as G3, and 20 (60.6%) were identified as mixed (G3G8 or G3G10) G types (Table 2). Additionally, 31 (93.9%) of the 33 P typed samples had the P[11] genotype and two (6.1%) exhibited dual P[1]P[11] types (Table 2). The majority of the mixed G types were G3G10 (55%, 11/20) following G3G8 (45%, 9/20) types.
The distribution of the G types showed that 52.9% were G3 and 47% were mixed G types. Among these, 35.3% were G3G8 (6/17) and 11.8% G3G10 (2/17) in the temperate region of western Himalaya while 25% of G3 and 75% mixed G types were found in the tropical, subtropical, and semi-arid regions. Among the mixed types, the most commonly detected G combination was G3G10 (55%). P types found in the temperate region of western Himalaya were mostly P[11] (94.1%) while 5.9% were the dual P type (P[1] P[11]). In the tropical, sub-tropical, and semi-arid regions (Table 2), 93.8% (15/16) were P[11] type and 11.1% (1/16) were the dual P type (P[1] P[11]).
The multiplex nested PCR typing results were further confirmed with sequencing, BLAST, and automated genotyping RotaC software analysis of selected isolates. As shown in Fig. 3, VP7 coding region sequence analysis of the bovine G3 isolates (B-46, 0970 and BR-133) showed that the predicted amino acid sequence identities with other bovine G3 RVA strains were between 90.5% (Bo/UK/1984 strain UKg9P GQ225779) and 98.8% (Bo/Ind/2006/RUBV3 EF200549). Identity was lower with bovine strains from the UK (90.5% to 92%) while two other bovine G3 strains isolates previously obtained from two different parts of India showed higher identities of 96.3% (J63 strain from central India) to 100% (RUBV3 strain from eastern India). Sequence identities of these three G3 isolates were between 89.9% and 95.1% with feline and canine G3 isolates, 93.3% and 94.5% with simian isolates, and 91.7% and 93.1% with equine isolates from Greece, Japan, and Germany. Equine G3 isolates from India had higher identities of 93.9% to 98.8%. All the G3 bovine isolates showed the lowest identity with porcine strains (82.8% to 91.4%). Sequence identities of these bovine strains with human G3 isolates from different parts of the world were between 89% and 94.5% (Fig. 3).
UK bovine G3 RVs showed a maximum of 183 variations at the nucleotide level corresponding to changes in the amino acid sequence at 26 positions compared to B-46, 0970, and BR-133 G3 strains. The RUBV3 strain from eastern India was more conserved as only one change was noticed at nucleotide level while there was no change at amino acid level. The J-63 G3 strain from central India showed 20 changes at the nucleotide level and eight changes at amino acid level (positions 22, 75, 146, 147, 211, 213, 221, and 242). Compared to the J-63 strain, the B-46 G3 strain isolated from the same location showed only 10 variations at the nucleotide level and four changes (59, 71, 76, and 317) at amino acid level.
Phylogenetic analysis of the B-46, 0970, and BR-133 G3 strains demonstrated that these three bovine strains clustered with RUBV3 and J-63 (bovine) and Erv-80 (equine). This finding indicated the genetic proximity of Indian bovine G3 strains to equine RVs (Fig. 3). On the other hand, other bovine RVA G3 strains from the UK (CP-1, PP-1, and UKg9P) clustered with human, canine, feline, and porcine G3 strains (Fig. 3).
Bovine RVA are associated with high mortality and morbidity among young calves, leading to substantial economic loss to livestock industry throughout the world, particularly in developing countries like India. Previous studies from India have shown a predominance of G10 in contrast to other G types (G6 or G8) among diarrhoeic calves [14,20,22,27,28,34]. However, a report from eastern India found a predominance of G6 rather than G10 as G6 was detected in 89.2% cases and G10 only in 4.6% cases [11]. Two new genotypes, G3 (3%) and G15 (2.3%), were also discovered in bovine calves in this study. Among the P types, P[11] was predominant (94.6%) followed by P[3] (3%), P[21] (1.5%), and P[14] (0.7%). This group also detected two unusual combinations, G15P[11] (1/130) and G15P[21] (2/130), in the calves [10]. Prior to these reports, Varshney et al. [33] discovered G3 types in diarrhoeic calves (10.7%) from central and southern India. The frequency of G3 was higher than the G6 or G8 genotypes and was second to the predominant G10 type. All these studies indicated that G type distribution varies according to geographical region. Similar patterns among bovine RV G6 populations have been observed in south Ireland [2].
G3 RV has been detected in canines, pigs, equines, and lapines. Unlike G6, G8, or G10 strains, G3 are rare in bovine calves [25]. To date, only two bovine RVA G3 strains from India (J-63 and RUBV3) and three from the UK have been characterized [5,11,25]. In the present study, the most prevalent genotypes were G3P[11] and mixed genotypes G3+G10P[11], G3G8P[11] and G3G8 P[1]P[11] and G3G10 P[1]P[11], which is uncommon for bovines. This indicates a changing pattern of genotype distribution from G6, G8, and G10 to a single G3 type or G8/G10 in combination with G3 as G3G8 or G3G10 among Indian bovine populations. Occurrence of the G3 genotype in central India was also recorded earlier by Varshney et al. [33], which further indicates that the G3 strain is continuously maintained in this region and might have spread to other parts of the country given that J-63 and RUBV3 are very close in genetic proximity. Variations at nucleotide and amino acid levels showed that J-63 and B-46 are from the same region but are genetically distinct.
Observations from the present study are in contrast to ones from previous reports [11,14,20,22,27,28,33,34] on the predominance of G10 and G6. During the present study none of the isolate was typed as G6 while a number of isolates were genotyped as G10 type but detected only in combination of G3 type. Genotyping failures in RVs has been reported earlier due to mismatching/mis-priming [1,2,24]; however, all the 33 RV isolates obtained during the present study were typable using genotyping PCR primers published by Gouvea et al. [12,13], Iturriza-Gómara et al. [16] and Taniguchi et al. [31]. The result confirms that no other genotype is circulating in the regions surveyed during the present study and strains are genetically more stable with no variations in the primer binding region that normally leads to failures in genotyping [1,2,24].
The results of present study clearly indicate that G3 in alone or combination with G10/G8 is predominant among bovine populations in India and is now the most prevalent genotype in many parts of the country. No early report from India identified combinations of G3 with G8 or G10. This could be either due to the use of only G6, G8, and G10 typing primers (the predominant G types) or re-assortment/interspecies transmission, which may have occurred more recently in bovine populations. Atypical detection of G3 or mixed G3 types in bovine populations indicates a complex interplay between the virus, host, and environment. Further studies to examine these relationships are warranted.
Comparison of the predicted amino acid sequences for B-46, 0970, and BR-133 G3 RVs to the sequences of other VP7 G3 strains showed that Indian bovine G3 strains are more conserved in comparison to UK bovine strains. The Indian bovine G3 strains appear to represent a re-assortment of equine (Erv80) and more recently emerged bovine G3 strains. These findings stress the need to further investigate any change in genotype distribution and the emergence of unusual genotypes through sequence analysis.
In conclusion, detection of G3 or mixed G3G10/G3G8 types in bovine populations provided significant information about the increasing circulation of G3 RV genotype in central and northern parts of India. Sequence analysis of the G3 strains uncovered evidence of multiple species re-assortment events. We observed changing patterns of bovine RVA genotypes with increased incidences of mixed genotypes containing G3 types. Our findings contribute to the growing body of evidence suggesting that interspecies transmission and re-assortment events between RVs can occur in nature. Therefore, examining the distribution of G and P types among bovine populations is important for understanding RV ecology and the mechanisms by which they evolve, cross the species barrier, exchange genes during re-assortment, and mutate via the accumulation of single point mutations or genetic recombination. Development of novel vaccines to effectively combat bovine diarrhoea due to RV will require incorporation of the emerging G3 types.
Figures and Tables
 | Fig. 1Diagrammatic illustration of VP7 (G)- and VP4 (P)-specific primer positions and type-specific amplicons. |
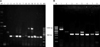 | Fig. 2Agarose gel electrophoresis showing amplicon sizes for the different G and P types. (A) G types, Lane M1: 1-kb DNA ladder, Lane M2: 100-bp DNA ladder, Lane 1: VP7 full-length amplicon (1062 bp), Lane 2: genotype G6 (NCDV strain), Lanes 3~6 and 11~13: G3 types, Lanes 7 and 8: G3G10 types (single sample showing amplification of two viz. G3 and G10 genotypes), Lanes 9 and 10: G3G8 types. (B) P types, Lane M: 100-bp DNA ladder, Lane 1: P[5] (UK strain), Lane 2: P[1] (NCDV strain), Lanes 3, 4, 6 and 7: P[11] type, Lanes 5 and 8: P[1]P[11] dual P types. |
 | Fig. 3Phylogenetic analysis of predicted VP7 amino acid sequences of bovine B-46, 0970, and BR-133 (G3 strains) with representative bovine G3 strains as well as other human and animal G3 strains. The tree was rotoed with Hu India 2009 116E3D (AB081593) RV strain sequences. The positions of bovine B-46, 0970, and BR-133 are indcated with black circles (•). The percent VP7 nucleotide and amino acid identities of the bovine B-46, 0970, and BR-133 strains with representative bovine G3 strains as well as other human and animal G3 strains are also presented. |
Acknowledgments
The authors wish to thank Prof. M.C. Sharma, the Director, Indian Veterinary Research Institute (IVRI), India, for conferring the project grant (No. 7-22/JDR).
References
1. Aladin F, Nawaz S, Iturriza-Gómara M, Gray J. Identification of G8 rotavirus strains determined as G12 by rotavirus genotyping PCR: updating the current genotyping methods. J Clin Virol. 2010. 47:340–344.

2. Cashman O, Lennon G, Sleator RD, Power E, Fanning S, O'Shea H. Changing profile of the bovine rotavirus G6 population in the south of Ireland from 2002 to 2009. Vet Microbiol. 2010. 146:238–244.

3. Collins PJ, Martella V, Buonavoglia C, O'Shea H. Identification of a G2-like porcine rotavirus bearing a novel VP4 type, P[32]. Vet Res. 2010. 41:73.

4. Das BK, Gentsch JR, Cicirello HG, Woods PA, Gupta A, Ramachandran M, Kumar R, Bhan MK, Glass RI. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol. 1994. 32:1820–1822.

5. El-Attar L, Dhaliwal W, Iturriza-Gómara M, Bridger JC. Identification and molecular characterization of a bovine G3 rotavirus which causes age-independent diarrhea in cattle. J Clin Microbiol. 2002. 40:937–942.

6. Esona MD, Mijatovic-Rustempasic S, Conrardy C, Tong S, Kuzmin IV, Agwanda B, Breiman RF, Banyai K, Niezgoda M, Rupprecht CE, Gentsch JR, Bowen MD. Reassortant group A rotavirus from straw-colored fruit bat (Eidolon helvum). Emerg Infect Dis. 2010. 16:1844–1852.

7. Estes MK, Kapikian AZ. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Rotaviruses. Fields' Virology. 2007. Vol. 2:5th ed. Philadelphia: Lippincott Williams & Wilkins;1917–1974.
8. Fagiolo A, Roncoroni C, Lai O, Borghese A. Borghese A, editor. Buffalo pathologies. Buffalo Production and Research. 2005. Rome: FAO;249–296.
9. Fukai K, Maeda Y, Fujimoto K, Itou T, Sakai T. Changes in the prevalence of rotavirus G and P types in diarrheic calves from the Kagoshima prefecture in Japan. Vet Microbiol. 2002. 86:343–349.

10. Ghosh S, Samajdar S, Sinha M, Kobayashi N, Taniguchi K, Naik TN. Molecular characterization of rare bovine group A rotavirus G15P[11] and G15P[21] strains from eastern India: identification of simian SA11-like VP6 genes in G15P[21] strains. Virus Genes. 2008. 37:241–249.

11. Ghosh S, Varghese V, Samajdar S, Sinha M, Kobayashi N, Naik TN. Molecular characterization of bovine group A rotavirus G3P[3] strains. Arch Virol. 2007. 152:1935–1940.

12. Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, Fang ZY. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990. 28:276–282.

13. Gouvea V, Santos N, Timenetsky Mdo C. Identification of bovine and porcine rotavirus G types by PCR. J Clin Microbiol. 1994. 32:1338–1340.

14. Gulati BR, Nakagomi O, Koshimura Y, Nakagomi T, Pandey R. Relative frequencies of G and P types among rotaviruses from Indian diarrheic cow and buffalo calves. J Clin Microbiol. 1999. 37:2074–2076.

15. Isegawa Y, Nakagomi O, Nakagomi T, Ishida S, Uesugi S, Ueda S. Determination of bovine rotavirus G and P serotypes by polymerase chain reaction. Mol Cell Probes. 1993. 7:277–284.

16. Iturriza-Gómara M, Kang G, Gray J. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J Clin Virol. 2004. 31:259–265.

17. Kapikian AZ, Hoshino Y, Chanock RM, Pérez-Schael I. Efficacy of a quadrivalent rhesus rotavirus-based human rotavirus vaccine aimed at preventing severe rotavirus diarrhea in infants and young children. J Infect Dis. 1996. 174:Suppl 1. S65–S72.

18. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970. 227:680–685.

19. Maes P, Matthijnssens J, Rahman M, Van Ranst M. RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol. 2009. 9:238.

20. Manuja BK, Prasad M, Manuja A, Gulati BR, Prasad G. A novel genomic constellation (G10P[3]) of group A rotavirus detected from buffalo calves in northern India. Virus Res. 2008. 138:36–42.

21. Matthijnssens J, Rahman M, Ciarlet M, Zeller M, Heylen E, Nakagomi T, Uchida R, Hassan Z, Azim T, Nakagomi O, Van Ranst M. Reassortment of human rotavirus gene segments into G11 rotavirus strains. Emerg Infect Dis. 2010. 16:625–630.

22. Minakshi , Prasad G, Malik Y, Pandey R. G and P genotyping of bovine group A rotaviruses in faecal samples of diarrhoeic calves by DIG-labelled probes. Indian J Biotechnol. 2005. 4:93–99.
23. Okada N, Matsumoto Y. Bovine rotavirus G and P types and sequence analysis of the VP7 gene of two G8 bovine rotaviruses from Japan. Vet Microbiol. 2002. 84:297–305.

24. Parra GI, Espinola EE. Nucleotide mismatches between the VP7 gene and the primer are associated with genotyping failure of a specific lineage from G1 rotavirus strains. Virol J. 2006. 3:35.
25. Rao CD, Gowda K, Reddy BSY. Sequence analysis of VP4 and VP7 genes of nontypeable strains identifies a new pair of outer capsid proteins representing novel P and G genotypes in bovine rotaviruses. Virology. 2000. 276:104–113.

26. Reidy N, Lennon G, Fanning S, Power E, O'Shea H. Molecular characterization and analysis of bovine rotavirus strains circulating in Ireland 2002-2004. Vet Microbiol. 2006. 117:242–247.

27. Saravanan M, Parthiban M, Ramadass P. Genotyping of rotavirus of neonatal calves by nested-multiplex PCR in India. Vet Arhiv. 2006. 76:497–505.
28. Sharma G, Taku A, Chhabra R, Bhat MA. Prevalence of bovine rotaviral diarrhoea in Jammu. Indian J Virol. 2009. 20:53–58.
29. Svensson L, Uhnoo I, Grandien M, Wadell G. Molecular epidemiology of rotavirus infections in Uppsala, Sweden, 1981: disappearance of a predominant electropherotype. J Med Virol. 1986. 18:101–111.

30. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007. 24:1596–1599.

31. Taniguchi K, Wakasugi F, Pongsuwanna Y, Urasawa T, Ukae S, Chiba S, Urasawa S. Identification of human and bovine rotavirus serotypes by polymerase chain reaction. Epidemiol Infect. 1992. 109:303–312.

32. Ursu K, Kisfali P, Rigó D, Ivanics E, Erdélyi K, Dán A, Melegh B, Martella V, Bányai K. Molecular analysis of the VP7 gene of pheasant rotaviruses identifies a new genotype, designated G23. Arch Virol. 2009. 154:1365–1369.

33. Varshney B, Jagannath MR, Vethanayagam RR, Kodhandharaman S, Jagannath HV, Gowda K, Singh DK, Rao CD. Prevalence of, and antigenic variation in, serotype G10 rotaviruses and detection of serotype G3 strains in diarrheic calves: implications for the origin of G10P11 or P11 type reassortant asymptomatic strains in newborn children in India. Arch Virol. 2002. 147:143–165.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download