Introduction
Streptococcus (S.) dysgalactiae subsp. equisimilis are beta haemolytic streptococci belonging to the Lancefield group C. Their normal habitat in the horse appears to be the skin and mucous membrane, though their isolation from aborted placentas and from abscessed lymph nodes suggest a possible pathogenic role [16]. Recently, the interest in these bacteria has increased because of their isolation from cases of strangles-like disease in absence of Streptococcusequi infection [10,16]. Furthermore, S. equisimilis infections in humans are increasing and are responsible for different clinical syndromes, including primary bacteraemia, pneumonia, endocarditis, arthritis, and streptococcal toxic shock syndrome [2,5,9,14].
In veterinary medicine, retrospective examination of bibliographic data about S. equisimilis infection is complicated by the frequent nomenclature changes due to the animal origin of the strains. At the beginning, the name S. equisimilis was proposed for beta-haemolytic C-streptococci frequently isolated from humans and thought to be uncommon in domestic animals. Further studies resulted in including these bacteria in the species S. dysgalactiae, while the name S. equisimilis lost its official value because it was not included in the new classification [17]. The two most recent taxonomic studies [17,18] were concordant in distinguishing two different subspecies of S. dysgalactiae, called S. dysgalactiae subsp. dysgalactiae and S. dysgalactiae subsp. equisimilis, although they were discordant about the strains to be included in the two subspecies. Consequently, on the basis of these studies, within a few years the name of the animal isolates changed from the original S. dysgalactiae, first into S. dysgalactiae subsp. dysgalactiae subsp. nov., and then into S. dysgalactiae subsp. equisimilis. Nevertheless, invalid or obsolete names continued to be used in the meantime.
S. equisimilis has been isolated infrequently from placentas from aborted, stillborn, and premature foals [6]. Recently, it has been isolated from horses with a history of respiratory disease or strangles-like disease [10]. However, infection in horses continues to be considered infrequent and opportunistic [16], though exhaustive epidemiological studies are not available and routine diagnostic tests are usually directed to identify other streptococci rather than S. equisimilis. Endocarditis and arthritis due to S. equisimilis infection have been recently described in swine [8], while the data published before refer to isolation of S. dysgalactiae without subspecies distinction [7]. Similarly, subspecies identification has not been carried out in cases of S. dysgalactiae infection associated with high mortality in amberjack (Seriola dumerili) and yellowtail (Seriola quinqueradiata) [13], and with polyarthritis in dairy goats [3]. In summary, exhaustive epidemiological data about S. equisimilis infection in animals are not available and, despite the fact that it is considered by some authors as an opportunistic pathogen, specific and definitive studies have been not carried out. Furthermore, considering the lack of information, it is also difficult to evaluate if the animals play a role in the maintenance and transmission of this potential zoonotic agent.
A PCR protocol to detect a common sequence of the species S. dysgalactiae in bovine milk has been published [15], and an in situ-PCR protocol for the detection of the subspecies equisimilis has been previously described in human formalin-fixed pulmonary samples from medicolegal autopsy cases [12]. In this latter case, the primers were specific for the streptokinase precursor gene and designed off the sequence of the equine isolate S. equisimilis 87-542-W. Considering the differences in the sequence of this gene among strains isolated from human, pig and horse [4], these primers are probably better suitable on equine samples because their high homology with the sequence of the equine strain 87-542-W. However, no data on PCR protocols were available to directly detect S. equisimilis in equine samples.
The aim of this study was to verify the reliability of a PCR protocol for rapid detection and identification of S. equisimilis both in streptococcal colonies and in equine nasopharyngeal swab samples in order to develop a diagnostic protocol for future epidemiological and pathogenetic studies. The evaluation of the test was based on a blind comparison among PCR and bacteriological results.
Materials and Methods
Samples and experimental design
A total of 99 monolateral nasal swab samples were collected from horses with a history of respiratory diseases for bacteriological and PCR investigations. Since S. equisimilis infections are considered to be infrequent and since the aim of this study was not to carry out an epidemiological survey, the samples were collected from horses from stud farms with a history of previous S. equisimilis infection in order to detect as many S. equisimilis positive swabs as possible. The nasal swab samples were briefly incubated in enrichment broth, as described below. After incubation, a loop of the enrichment broth was spread on blood agar for bacterial culture, and 1 mL of the remaining broth was used for DNA extraction and PCR. By this method it has been possible to use the same swab sample to test by both bacteriology and PCR instead of collecting two separate swabs.
Beta-hemolytic and catalase negative streptococci grown on sheep blood agar were used for the determination of the Lancefield group for biochemical identification and for PCR. Bacteriological and bio-molecular assays were carried out blindly by different operators in different laboratories, and comparison among the results was done only after both assays were completed.
Bacterial culture and biochemical identification
Swab samples were incubated in 4 mL T14 enrichment broth [phosphate buffered saline pH 7.2, 2% fetal calf serum (Celbio, Italy), 0.0005% amphotericin B (Bristol-Myers Squibb, Italy)] at 37℃ for 6 h and then spread on agar plates containing 5% sheep blood and Streptococcus selective supplement (Oxoid, Italy). Beta-hemolytic and catalase negative colonies were subcultured for purity, processed for Lancefield group classification by latex agglutination test (Slidex Strepto-Kit; BioMerieux, Italy), and identified by their biochemical profile using API 20 Strep (BioMerieux, Italy) following the manufacturer instructions. A suspension of S. equisimilis ATCC 10009 was used as positive control for each batch of API 20 Strep tests.
Template preparation for PCR assays
The reference strain of S. equisimilis ATCC 10009 and other 18 S. equisimilis strains previously isolated from horses in Central Italy and conserved in the laboratory collection (Laboratory of Medical Microbiology and Infectious Diseases - University of Camerino, Italy) were used to optimize DNA extraction and PCR protocols from colonies. Furthermore, a loop of a pure culture of the reference strain was dissolved in 4 mL of T14 enrichment broth, incubated at 37℃ for 6 h and used to optimize the DNA extraction protocol from broth samples.
DNA samples were prepared from colonies grown on blood agar and from 1 mL T14 enrichment broth after incubation at 37℃ for 6 h with nasal swab samples. Briefly, DNA extraction from colonies was carried out similarly as previously described with few modifications [1]. One loop of colonies from the pure culture was suspended in 50 µL digestion buffer (Tris-HCl 10 mmol/L, EDTA 1 mmol/L, pH 8.0 containing 5 U/µL lysozime; Sigma, Italy) and incubated at 37℃ for 30 min in a water bath. 0.75 µL proteinase K 20 µg/µL (Eurobio, France) were subsequently added, and the solution was incubated at 56℃ for 30 min. After boiling for 10 min, the samples were centrifuged at 10,844 × g for 5 min and the supernatant was transferred in a new tube and cooled before use.
Furthermore, 1 mL of T14 enrichment broth, after incubation with nasal swab, was centrifuged at 21,255 × g for 10 min, the supernatant was discarded, and the pellet was suspended in 25 µL digestion buffer at 37℃ for 60 min. After adding 0.38 µL proteinase K 20 µg/µL (Eurobio, France), the samples were boiled, centrifuged and transferred in new tubes as described above.
PCR protocol
Three separate PCR tests, based on published primers, were used to detect S. dysgalactiae and the subspecies equisimilis. For S. dysgalactiae detection, the primers Sdy519-F 5'-GGC TCA ACC ACT NTA CGC TT-3' and Sdy920-R 5'-ATC TCT AGA CCG GTC AGG A -3' were used for the amplification of a 401 bp sequence of the 16SrRNA region [15]. PCR was carried out in a Hybaid PCR Express Thermal Cycler (Hybaid, UK), and the PCR reaction mixture (25 µL) contained 12.5 µL of Taq PCR mastermix (Qiagen GmbH, Germany), 10 pmol of each primer and 2 µL of DNA template prepared from colonies or 5 µL of DNA template prepared from T14 enrichment broth. In order to verify the absence of inhibition or cross-contamination, S. equisimilis ATCC 10009 DNA or water were used in each PCR run instead of the template DNA respectively as positive and negative control samples. PCR was conducted with the following program: 94℃ for 3 min, 35 cycles at 94℃ for 30 sec, at 57℃ for 30 sec, and at 72℃ for 40 sec, followed by a final extension at 72℃ for 7 min.
For the detection of S. equisimilis, the primers eqsim-F 5'-TCA AAT CGG TTG GCA CAG AC-3' and eqsim-R 5'-CGT CCT TAG CAT AGA AGG ATT GG-3 flanking a 279 bp fragment of the streptokinase precursor gene were used [12]. The PCR was conducted as described above, but the annealing temperature was 55℃ instead of 57℃.
The presence of PCR products was determined by the electrophoresis of 10 µL reaction products in 1.5% agarose gel containing 0.5 µg/mL of ethidium bromide with Tris-borate-EDTA buffer (TBE; 89 mM Tris, 89 mM boric acid, 2 mM EDTA pH 8.3). A sample was considered positive for S. equisimilis when both Sdy and eqsim PCR were positive.
In parallel experiments, the Sdy and eqsim primers were used in a duplex PCR using the PCR mixture and the protocol described above. On the basis of preliminary experiments using DNA extracted from the reference strain of S. equisimilis, the annealing temperature for the multiplex PCR experiments was fixed in 57℃ and no additional MgCl2 was included in the mixture.
PCR detection limit
The detection limit of the PCR protocol for S. equisimilis amplification was determined using a series of 10-fold serial dilutions of DNA purified from S. equisimilis ATCC 10009 strain. The preparation of the template, PCR assays and visualization of the amplification products on agarose gel were carried out as above described for the tested samples. The starting DNA concentration was determined by spectrophotometer reading (Genova; Jenway, UK), and 10-fold dilutions were carried out in TE buffer pH 7.0 (1 M Tris-Cl, 0.5 M EDTA, pH 8.0).
PCR specificity
Although the primer sets to detect S. dysgalactiae spp. and the subspecies equisimilis have been previously published [12,15], their specificity was verified again in this study on different streptococci and equine respiratory bacteria obtained from the laboratory collection (Laboratory of Medical Microbiology and Infectious Diseases - University of Camerino, Italy). The cultures included Streptococcus equi subsp. equi (n = 5) (two of them kindly provided by Prof. J. Alber and Prof. C. Lämmler, Justus-Liebig-Universität of Gieβen, Germany), Streptococcus dysgalactiae subsp. dysgalactiae (n = 3), Streptococcus equi subsp. zooepidemicus (n = 5), Streptococcus mitis (n = 2), Streptococcus pneumoniae (n = 1), Streptococcus pyogenes (n = 4), Streptococcus canis (n = 3), Staphylococcus aureus (n = 4), Staphylococcus intermedius (n = 4), Rhodococcus equi (n = 5), Escherichia coli (n = 3), Klebsiella pneumoniae (n = 1), and Pseudomonas aeruginosa (n = 3). Furthermore, 40 nasal swab samples and 20 guttural pouches wash samples collected from 60 healthy horses that tested negative for S. equisimilis by bacteriology were used to verify the absence of non-specific PCR results.
Statistical analysis
The results from bacteriology and PCR assays were analyzed to assess the statistical agreement between the two methods. The kappa statistic was calculated as the observed agreement beyond chance divided by the maximum agreement beyond chance [11]. For convenience, a freeware computer program was used to perform the calculations (Kappa statistics calculator, version 02/07/2001 by Jen-Hsiang Chuang; Columbia University, USA). Values of kappa between 0.6 and 0.8 are indicative of substantial agreement, and values of kappa between 0.8 and 1.0 are indicative of almost perfect agreement [11].
Results
Bacteriological examination and biochemical identification
Beta-hemolytic and catalase negative streptococci of the Lancefield group C demonstrating the biochemical profile of S. equisimilis were isolated from 23 (23.23%) out of the 99 tested samples. In one case, the API 20 Strep test had to be repeated because an unacceptable profile was obtained after 24 hours of incubation.
PCR
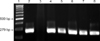
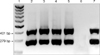
PCR using Sdy primers resulted in a 401 bp segment in the S. equisimilis ATCC 10009 strain, in the 18 S. equisimilis positive controls, and in 23 (23.23%) isolates and 29 (29.29%) broth samples out of the 99 equine nasal swabs collected. Products 279 bp in size were obtained from the same samples by PCR using eqsim primers (Fig. 1). No bands were obtained by testing the negative controls including the bacteria from the laboratory collection and the 40 nasal swabs and the 20 guttural pouches wash samples collected from the uninfected horses. Identical results were obtained by multiplex PCR using Sdy and eqsim primes (Fig. 2). The bands visualised on agarose gel were well defined and no aspecific products were observed. Running 10 µL of amplification products from DNA extracted from colonies often resulted in an overcharging of the wells, but it appeared useful for the visualisation of the PCR products from DNA extracted from broth samples, where running of a smaller amount of amplification products occasionally resulted in thin and weak bands (data not shown).
The detection limit of the eqsim primer set determined using a series of 10-fold dilutions of template DNA from the reference strain of S. equisimilis was 10 fg/reaction (Fig. 3).
Comparison between bacteriological and PCR results
Results of culture and PCR for S. equisimilis were concordant in 23 out of 23 samples (kappa = 1.00 - "perfect agreement"). Furthermore, PCR was positive not only in the 23 T14 enrichment broth samples that were positive by bacteriology, but also in 6 out of the 76 T14 enrichment broth samples that were negative for S. equisimilis by culture (kappa = 0.84 - "almost perfect agreement").
Discussion
S. equisimilis can infect humans and several animal species. Its pathogenic role has been well documented in humans, but no detailed data have been reported so far in horses. The availability of rapid and reliable diagnostic tools is fundamental for detecting S. equisimilis in clinical specimens and for its differentiation from other streptococci. Currently, bacterial culture and biochemical identification with commercial kits are widely used for S. equisimilis diagnosis. Nevertheless, overgrowing of other beta-haemolytic streptococci or mistakes in biochemical profile interpretation may be encountered during bacterial examination. PCR is widely used in many fields of microbiology, such as for direct bacterial detection in clinical specimens or for rapid identification of the cultured colonies. S. equisimilis secretes a plasminogen activator, known as streptokinase, which catalyses the conversion of plasminogen to plasmin, and may facilitate tissue invasion [4]. The nucleotide sequence of the streptokinase precursor gene of an equine strain of S. equisimilis has been sequenced and published [4] and may represent a target sequence for the binding of specific primers. Furthermore, a PCR protocol to detect a common sequence of the species S. dysgalactiae has been previously described [15] and may be used as a control for species identification.
The multiplex PCR protocol described in this study has been able to specifically detect both the streptokinase precursor gene of S. equisimilis and the 16SrRNA region of the species S. dysgalactiae in equine nasal swabs and in colonies grown on blood agar plates. There was "perfect agreement" (kappa = 1.00) between bacteriological examinations and PCR in identifying S. equisimilis colonies. The agreement between the two methods in detecting S. equisimilis in enrichment broth samples inoculated with equine nasal swabs was "almost perfect" (kappa = 0.84), and PCR protocols showed a higher sensitivity than bacteriology. All the samples where S. equisimilis was isolated by bacteriology were also positive by PCR. Furthermore, PCR was able to detect S. equisimilis DNA in 6 samples more than bacterial culture. This could be due to not only to the different characteristics of the two techniques, but also to the amounts of the starting samples examined by the two methods. The preliminary culture of the swabs in enrichment broth first causes multiplication, and consequently, a higher concentration of the bacteria. Nevertheless, only 100 µL of this solution are tested by standard bacteriological examination, while 10-fold of this volume (one-fourth of the total volume) was analysed by PCR in this study without difficulty, resulting in its higher sensitivity.
Bacterial culture is a fundamental tool for S. equisimilis diagnosis, but the PCR assay can identify S. equisimilis isolates or detect S. equisimilis DNA directly in clinical specimens. In this latter case, a preliminary incubation in enrichment broth is recommended to increase the DNA yield. The PCR technique may be particularly useful when sample conservation does not guarantee the viability of the bacteria or when the samples have been treated with bacterial killing substances, such as ethanol or formalin, for other experiments (i.e. histology or cytology). In any case, the contemporary evaluation of diagnostic samples by bacterial culture and PCR could be combined for higher diagnostic sensitivity and specificity with the availability of live bacteria.
In conclusion, this study describes a rapid duplex PCR protocol for the detection and identification of S. equisimilis in equine nasal swabs by amplification of the streptokinase precursor gene and by contemporaneous amplification of a 16SrRNA region specific for the species S. dysgalactiae. This method could be useful for rapid diagnosis and epidemiological purposes by the direct detection of S. equisimilis in equine nasal swabs.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download