Haemophilus (H.) parasuis is the causative agent of Glässer's disease, which has caused serious problems in the pig industry worldwide [11]. A total of 15 serovars of H. parasuis have been defined, and some strains are cannot be typed by serotyping [5,14]. This serovar diversity and lack of extensive cross-reaction between serovars makes it difficult to diagnose and control H. parasuis infections [10]. Several molecular methods, including PCR [9], in-situ hybridization [4], improved species specific PCR [1] and real-time PCR [15], have been developed for rapid detection of H. parasuis. In addition, some serological methods have also been developed for antibody detection after H. parasuis infection, including the indirect hemagglutination test (IHA) [6], enzyme-linked immunoassay [6] and the complement fixation test [7]. However, a universal serological method for detecting this pathogen has not been developed to date [12]. Despite the lack of extensive cross-reaction between serovars, weak cross-reactions were still observed between some strains with different serovars [3,10,14]. This study was conducted to develop a universal plate-agglutination test for detection of H. parasuis by mixing the antisera of serovars as diagnostic antisera.
Reference strains representing serovars 1 through 15 of H. parasuis were kindly provided by the Queensland's Animal Research Institute of Australia. Seventy five clinical isolates of H. parasuis were isolated from clinical swine cases of meningitis, arthritis or pneumonia in swine suffering from Glässer's disease in China from 2006 to 2008. All clinical isolates were identified by PCR [9] and serotyped by the IHA test [14]. A total of 22 other bacterial strains covering 12 species were used for specificity evaluation. Table 1 shows the details of the strains used in this study.
Strains of H. parasuis were grown on tryptone agar (BeiJing AoBoXing Universeen Bio-Tech, China) supplemented with 0.02% (w/v) NAD (Sigma, USA) and 5% newborn calf serum (Shanghai Shell Gene Biotech, China) at 37℃ under 5% CO2. Antisera against reference strains representing serovars 4, 5, 12, 13 and 14 were prepared as described previously [5,13]. Briefly, strains grown overnight were harvested with phosphate-buffered saline solution (pH 6.8) containing 0.3% formalin and then kept at 37℃ for 18 h. Formalinized-whole-cell (FWC) suspensions were adjusted to a concentration of 5.0 × 109 cells per ml. Two ml of the suspensions and an equal volume of Freund's incomplete adjuvant were injected subcutaneously at four sites. Three weeks later, rabbits were given an intravenous inoculation of 0.5 mL of FWC suspension followed by six doses administered intravenously in increasing doses twice a week. Antisera were separated seven days after the last injection.
All of the antisera were absorbed with an equal volume of whole cells mixture of 10% Salmonella and Escherichia (E.) coli. Hyperimmune antisera were diluted with 0.85% NaCl containing 1 : 10,000 thimerosol to the titer of 1 : 200 in the pate-agglutination test, after which they were stored at -20℃ until further analysis. Diagnostic antisera were prepared by mixing antisera of serovars 4, 5, 12, 13 and 14 in the optimized ratio of 1 : 1 : 1 : 2 : 1, which was determined by orthogonal experiments for the plate-agglutination test.
To observe the cross-agglutination between serovars, antisera of serovars 4, 5, 12, 13, 14 and the diagnostic antisera were used to react with 15 reference strains in a plate-agglutination test.
A total of 75 H. parasuis clinical isolates were used to validate this method. To evaluate both the repeatability and reproducibility, three parallel experiments were conducted for each sample. To determine the specificity of the diagnostic antisera, a total of 22 non-H. parasuis strains covering 12 species (Table 1) were tested in the same method as described above.
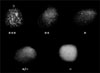
A drop of the diagnostic antisera (approximately, 10 µL) was placed on a clean glass surface, after which a small amount of an H. parasuis colony was removed from the culture media with an inoculating needle and mixed with the antisera. All reactions were conducted at room temperature. For the purposes of this study, a five-point scale (exhibited pictorially in Fig. 1) was adopted and defined as follows: heavy flocculent precipitates (+++) forming within 10 sec (clear background); heavy flocculent precipitates (++) taking 10 to 20 sec to form (clear background); light flocculent precipitates (+) taking 10 to 20 sec to form against a mostly clear background; light flocculent precipitate (+/-) against a cloudy homogeneous background after extended incubation (20 sec to 30 sec); negative reactions (-) remained turbid against a cloudy homogeneous background.
When the antisera against serovars 4, 5, 12, 13, an 14 were reacted with the 15 reference strains, cross-reactions were observed between some serovars; however, extensive cross reactions were not observed between serovars (Table 2). Interestingly, the diagnostic antisera combined with the antisera of serovars 4, 5, 12, 13 and 14 could agglutinate with all 15 reference strains of H. parasuis.
A total of 75 H. parasuis clinical isolates were detected in this study. Except for a non-typeable isolate giving a negative result, 74 of 75 clinical isolates were found to be positive by the plate-agglutination test (Table 1). The results of three independent tests were identical; therefore, this serological method showed good repeatability and reproducibility. None of the related species reacted with the diagnostic antisera in this study (Table 1), indicating that the diagnostic antisera could differentiate H. parasuis from the non-H. parasuis strains.
Epidemiological studies of H. parasuis infections showed that serovars 4, 5, 12, 13, 14 and non-typeable isolates were most prevalent among isolates from field cases worldwide [2,4,5,10,11]. In this study, a universal plate-agglutination test that could detect H. parasuis of 15 serovars and nearly all non-typeable strains was developed by preparing polyvalent antisera with the antisera of five prevalent serovars. The results of this test were highly consistent with the PCR technique developed by Oliveira et al. [9] and the test requires no special antigen preparation. In our preliminary test, the antisera of H. parasuis could agglutinate with Salmonella and E. coli. The cross-reaction could be eliminated after the antisera were absorbed with the whole cells of Salmonella and E. coli. The plate-agglutination test is a rapid, simple and useful method for detecting H. parasuis species directly from culture medium, and is an accessible serological test that can be conducted in any diagnostic laboratory.
H. parasuis strains, including virulent and non-virulent strains, are normally found colonizing the nasal cavity and tonsils of conventional pigs and, to a lesser extent, of healthy animals. Consequently, samples obtained from the upper respiratory tract should not be used to assess H. parasuis infections in the herd by PCR [9]. In agreement with the PCR assay described above, the plate-agglutination test developed in this study cannot distinguish virulent strains from non-virulent strains. Accordingly, samples obtained from systemic sites, such as pleura, pericardium, peritoneum, meninges or joints, are better options for diagnosis of H. parasuis infections with the plate-agglutination test.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download