Porcine endogenous retrovirus (PERV) is a member of the family Retroviridae, genus Gamma retrovirus. Based on the sequences of envelope genes, PERV has been classified into 3 subtypes, PERV-A, -B, and -C [8]. Many PCR abased PERV detection techniques have been developed, including conventional PCR, RT-PCR [6,7] and quantitative real-time PCR [1]. The dual priming oligonucleotide (DPO) system is an innovative technique that can reduce non-specific amplification during PCR. Thus, multiplex PCR using DPO based primers could enhance the specificity and sensitivity of detection of several targets simultaneously [2]. The objective of the current study was three-fold: to develop a DPO based multiplex PCR assay; to apply the developed assay to field samples for the detection of PERV subtypes in tissue and blood samples; and to compare hair root and tissue samples as source material for the detection of PERV proviral genes.
Primer sets were designed using the design principles of the DPO system, as previously described [2,5]. The sequences of the primers are listed in Table 1. DNA extraction was performed in accordance with a previous study [7]. For PERV subtyping, 1 µL of porcine genomic DNA and 500 nM of each primer mixture were added to AccuPower HotStart PCR Premix (Bioneer, Korea). The following thermal cycling parameters were used for PCR: 40 cycles of denaturation at 94℃ for 30 sec, annealing at 56℃ for 30 sec, and extension at 72℃ for 50 sec. Upon completion, samples were maintained at 72℃ for 5 min prior to cooling.
The specificity of the primers was tested using the following cell lines: Madin-Darby bovine kidney (MDBK), Madin-Darby canine kidney (MDCK), Vero, porcine kidney (PK)-15, human rectal tumor (HRT)-18, neuro 2a (N2a) and primary chicken embryonic fibroblasts (CEFs). Extracted DNA was reconstituted at a concentration of 10 ng/µL. The template DNA used as positive control in this study was from miniature pigs because the PK-15 cell do not have the envC gene [3]. To evaluate the specificity, the amplified PCR products were sequenced (Genotech, Korea), and the sequence data was analyzed using MegAlign (DNAStar, USA) with reference sequences.
To determine the sensitivity of the assay, genomic DNA was extracted from the liver of a miniature pig and used as a positive control. The concentration of DNA was measured, and then adjusted to a concentration of 10 ng/µL. Five 10-fold serial dilutions of DNA were prepared for sensitivity test. Comparison of the multiplex and single PCR assay was performed using tissue and blood samples. Liver samples from 20 purebred Duroc and 20 miniature pigs were obtained for the test on tissue samples. The pigs were divided into 4 age groups (10 days old, 30 days old, 70 days old, 110 days old) containing 5 pigs each. Blood samples which were treated with EDTA were obtained from 8 Yorkshire, 12 Landrace, and 20 mixed breed (Landrace×Yorkshire) sows. Liver tissue (1 g) was homogenized in 1 mL of phosphate buffered saline and buffy coat was collected from blood samples. The single PCR with primers for envelope genes for PERV proviral DNA established in our previous study [7] was employed. In order to compare the detection efficiency of PERV subtypes between the newly developed multiplex PCR and the previous single PCR, PERV pol gene and pig mitochondrial DNA were omitted from the current research. The conditions for the single PCR were the same as for the multiplex PCR.
All animal experiments were carried out in accordance with the current laws of Korea. The care and treatment of the animals was done in accordance with the protocols and guidelines of the Seoul National University Institutional Animal Care and Use Committee.
To compare the efficiency of the multiplex PCR technique to detect the PERV proviral genes pol, envA, envB and envC in hair root and tissue samples from the same individual, genomic DNA was extracted from hair and liver samples from 16 deceased pigs that had been submitted to the Veterinary Virology Laboratory, College of Veterinary Medicine, Seoul National University for disease diagnosis. DNA extraction from hair was from 15 strands of hair including roots in accordance with a previous study [4]. The detection of PERV proviral genes in hair and liver was carried out using multiplex PCR, and compared.
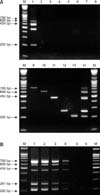
There was no evidence of amplification of target sequences from HRT-18, N2a, MDBK, MDCK, CEFs and Vero cells, or from distilled water (Fig. 1A). Expected amplification was only generated in samples of pig origin. The nucleotide similarity among amplified sequences and reference strains was 99% for PERV pol, envA, envB, envC, and 100% for porcine mitochondrial DNA. Likewise, the sensitivity of the multiplex PCR was demonstrated by simultaneous amplification of all target genes in as little as 10 pg of genomic DNA (Fig. 1B). All the tissues from 20 Duroc and 20 miniature pigs and blood samples from 40 three breed of pigs were positive for PERV pol and mitochondrial DNA. All of the tested liver tissue samples were positive for envA and envB proviral genes in both single and multiplex PCR methods. PERV envC was detected in all of the samples tested from miniature pigs by both methods, whereas only 9 and 12 samples from 20 Duroc pig samples were positive for PERV envC in single PCR and developed DPO based multiplex PCR, respectively. Although the tested Duroc pig samples from purebred, the existence of envC proviral DNA differed by individual. The existence of envC proviral DNA might not be related to genetic constitutions but the characteristics of individuals as described in previous study [4] showing absence of envC within same bred. Further experiments based on the kinship of pigs and with long-term follow up would be helpful in producing more valuable data in this regard. All of the tested blood samples were positive for PERV envA and envB proviral genes by both methods. PERV-C proviral genes were detected in 27 and 32 samples from 40 blood samples by single PCR and multiplex PCR, respectively. Therefore, developed DPO based multiplex PCR detected more samples which were positive for envC proviral gene compared to single PCR, established previously [7]. Although the primer designed in this study might not be optimal for detection of certain PERV-C variants, the improvement of the technique has been verified by in this manuscript by comparing with our previous study [7]. We also compared the ability of the multiplex PCR assay to distinguish PERV subtypes from hair and liver tissue samples using 16 conventional pigs. The pattern and subtype of PERV proviral genes in hair root samples was identical to that of liver samples. Of 16 commercial animals tested, all were positive for PERV pol, envA and envB, while only 6 were positive for envC in both hair and liver samples. These results might provide strong support for the use of hair in screening and subtyping of PERV, which eliminates the need to sacrifice the animal. Multiplex PCR or RT-PCR assays using DPO-based primers have been previously developed for the simultaneous detection of several species of virus [5]. To date, there have been no reports of PERV subtyping using multiplex PCR. Thus, the current study is the first attempt to detect PERV subtypes using DPO-based primers.
The multiplex PCR assay using DPO-based primers was developed and successfully applied for the analysis of field samples. In conclusion, this multiplex PCR method is a rapid, economic screening assay for detection of the PERV subtypes in pig or pig originated materials.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download