Abstract
Bacterial cold water disease, enteric red mouth disease and frunculosis are the common bacterial diseases of fish worldwide. The etiologic agents of these diseases are Flavobacterium (F.) psychrophilum, Yersinia (Y.) ruckeri and Aeromonas (A.) salmonicida subsp. salmonicida, respectively. In this study, a multiplex polymerase chain reaction (m-PCR) method with YER8/10-Fer3/4-FP1/3 primer pairs which can identify these fish pathogens simultaneously was developed and optimized. In optimized conditions, neither false specific nor nonspecific amplification occurred. The detection limits of the m-PCR method using DNA extracts from dilutions of pure cultures of bacteria were 35 pg for Y. ruckeri and F. psychrophilum and 70 pg for A. salmonicida subsp. salmonicida. It was determined that 15 CFU Y. ruckeri and F. psychrophilum and 30 CFU A. salmonicida subsp. salmonicida could be detected by m-PCR developed using genomic DNA extracted from dilutions of the suspensions. The detection limits in the presence of tissue debris were 125 CFU for Y. ruckeri and F. psychrophilum and 250 CFU for A. salmonicida subsp. salmonicida. In conclusion, we submit that the m-PCR method developed and optimized in this study can be used for accurate and rapid identification of these bacteria.
Aquaculture is an emerging industrial sector that requires more research with scientific developments and innovation. Under intensive aquaculture conditions, the risk of stress increases and a significant proportion of stock may become infected. Healthy looking fish without any clinical signs or lesions can carry some pathogens and create a serous risk for spread of contagious diseases in fish populations. Bacterial cold water disease, enteric red mouth disease and frunculosis caused by Flavobacterium (F.) psychrophilum, Yersinia (Y.) ruckeri and Aeromonas (A.) salmonicida, respectively, are common major contagious diseases worldwide in culture fisheries of salmonids. These diseases also have significance in respect of considerable economic losses [1,3].
Flavobacterium psychrophilum, a fastidious Gram negative agent, causes cold water disease and affects mainly salmonids (Oncorhynhus mykiss) and coho salmon (Oncorhynhus kisutch). Although its cultivation from fish tissues is performed on agar media and identified using some biochemical and serological techniques, correct identification based on phenotypic characteristics is reported to be difficult and time consuming [4,17]. Polymerase chain reaction (PCR) has also been used to detect Flavobacterium species [4]. Enteric red mouth disease, caused by Y. ruckeri, is a septicemic disease occurring primarily in salmonids. However, there have also been reports of nonsalmonids being affected. Several stress factors are important determinants in the development of this infection. Clinically healthy fish may be carriers of this pathogen for transmission to other fish [12] and biochemical tests and immunological assays have been used for its identification. In addition, PCR can be used to detect Y. ruckeri in some internal organs and feces [2]. A. salmonicida causes Frunculosis, a disease characterized by muscle lesions and associated skin ulcers and septicemia. A. salmonicida is commonly isolated from both salmonids and nonsalmonids [9]. Different diagnostic methods such as ELISA, agglutination tests and PCR have been used to detect this infection [2,10,12].
Selective culture, standard biochemical methods, and serological and histological examinations have traditionally been used to detect and enumerate bacterial pathogens. These methods have some disadvantages such as low sensitivity, low specificity, tediousness and high time consumption [1,10]. Thus, it is obvious that a more cost effective, sensitive and specific system is required for detection of these fish pathogens. Such a system would also be useful for surveillance and monitoring fish populations. Recently, molecular biological methods such as PCR, restriction enzyme digestion and probe hybridization have improved diagnostic sensitivity and specificity. These methods allow more rapid and accurate identification of asymptomatic or carrier fish. More recently, the use of multiplex PCR (m-PCR) has provided rapid and highly sensitive methods for the specific detection of pathogenic microorganisms in the aquatic environment. Furthermore m-PCR assay is more rapid and cost-effective than simplex PCR [3,10,13]. However, while m-PCR has been shown to be a valuable method for the identification of several pathogens in the field of infectious diseases, it is not yet a common tool for detection of fish pathogens. In this work, we aimed to develop and optimize the m-PCR assay for the simultaneous detection of F. psychrophilum, Y. ruckeri and A. salmonicida from pure cultures and tissues.
Taxonomically related and unrelated bacterial strains used to evaluate m-PCR method in this study are presented in Table 1. Trypticase soy broth (TSB) and trypticase soy agar (TSA) were used for cultivation of Aeromonas spp. and Yersinia spp. For cultivation of Flavobacterium spp., enriched Anacker-Ordal agar (EAOs, 0.5% tryptone, 0.05% yeast extract, 0.02% sodium acetate, 0.02% beef extract, 5% horse serum with 1.5% agar, pH 7.2~7.4) was used [14]. Aeromonas salmonicida subsp. salmonicida was incubated at 17℃, A. hydrophila at 35℃, Y. ruckeri at 22~25℃, Y. enterocolitica at 35℃ for 24~48 h. F. psychrophilum and F. columnare were incubated for 3~5 days at 19℃ and 25℃, respectively. After incubation periods, two colonies grown on mediums were transferred to TSB with 15% glycerin and stored at -20℃ until use.
DNA extractions from strains were performed using a commercial extraction kit (Omega Bio-Tek, USA) based on filtration through a spin colon as per the manufacturer's recommendations.
The oligonucleotide primer sets used in the experiments were designated according to the references presented in Table 2. The oligonucleotides used for each microorganism in the M-PCR assay were synthesized by Fermentas (Lithuania). The expected size of PCR products with these primers are also presented in Table 2. Three experiments were performed to establish the specificity of the m-PCR assay using different combinations of these primer sets specific for Y. ruckeri, A. salmonicida subsp. salmonicida and F. psychrophilum. In the first experiment (Trial 1) YER8/10, PAAS1/2 and FP1/3 were used. Ruck1/2, PAAS1/2, FP1/3 primer sets and YER8/10, Fer3/4, FP1/3 primer sets were used in the second (Trial 2) and the third experiments (Trial 3), respectively. To evaluate the specificity of m-PCR assay, genomic DNA from three bacteria (A. salmonicida subsp. salmonicida ATCC 33658, F. psychrophilum DSMZ 3660 and Y. ruckeri ATCC 29473 strains) and some different taxonomically and/or ecologically related or unrelated bacteria (Table 1) were used.
In these experiments, 25 µL of PCR master mix was prepared containing DEPC-treated water, 1× PCR buffer, 2 mM of MgCl2, 0.2 mM of each dNTP, 1.5 U of Taq polymerase, 1 mM of each primer and 3 µL template DNA. The amplification was carried out under the following conditions: an initial denaturation step at 94℃ for 2 min, followed by 35 cycles of amplification (denaturation at 94℃ for 40 sec, annealing at 60℃ for 40 sec, and extension at 72℃ for 1 min) and a final elongation period at 72℃ for 5 min. These conditions were determined as an initial protocol for subsequent m-PCR experiments. The amplicons were transferred to 1.5~3% agarose gel and electrophoresed. DNA bands were stained with ethidium bromide (2 µg/mL) and visualized by a UV transilluminator.
For the evaluation of the sensitivity of m-PCR, the minimum detectable amounts of DNA in m-PCR were detected for A. salmonicida subsp. salmonicida ATCC 33658, F. psychrophilum DSMZ 3660 and Y. ruckeri ATCC 29473 strains. After extraction of DNA, the genomic DNA concentration of each strain was measured using a spectrophotometer and adjusted to 35 ng/mL. Serial dilutions of each DNA strain were prepared with sterile distilled water at concentrations ranging from 35 ng/mL to 1.7 pg/mL. Aliquots of 5 µL of each dilution of genomic DNA were mixed with the respective aliquots of the other two strains. These DNA mixtures were used as template DNA for the m-PCR experiments, and the detection limit for DNA was determined.
For the evaluation of sensitivity of m-PCR from the cultures, the logarithmic phase culture was prepared for each bacterium in an appropriate broth. To enumerate the stock cultures, serial ten-fold dilutions of pure culture suspensions of each bacterium were prepared in sterile 0.9% NaCl solution. Zero point one mL of each dilution was spread onto TSA for A. salmonicida subsp. salmonicida ATCC 33658 and Y. ruckeri ATCC 29473, EAOs for F. psychrophilum DSMZ 3660. Aeromonas salmonicida subsp. salmonicida and Y. ruckeri were incubated at 25℃. F. psychrophilum strain was incubated at 19℃. After incubation period, colonies were counted and bacterial concentration of stock cultures was detected as CFU/mL. Then, the concentration of the stock suspension of each bacterium was adjusted to 1.0 × 108 CFU/mL. The 5 serial ten-fold dilutions and followed by 7 serial two-fold dilutions of each bacterial stock suspension were prepared (1.0 × 107, 1.0 × 106, 1.0 × 105, 1.0 × 104, 1.0 × 103, 500, 250, 125, 62, 31, 15, 7 CFU/mL). One mL of each dilution was centrifuged at 10,000 rpm for 3 min. The supernatant was removed and 200 µL of sterile distilled water was added to the pellet. This suspension was mixed by a vortex and the mixture then boiled for 10 min and then centrifuged at 10,000 rpm for 9 min. The supernatant was used as template DNA. In m-PCR assays, each bacterial DNA from each dilution was used by mixing with the respective aliquots of other bacteria.
To determine the sensitivity of the m-PCR method in the presence of tissue debris, the liver from healthy salmon trout was homogenized with Tris-EDTA (TE) buffer (1 mM Tris-HCl, 0.5 mM EDTA [pH 8]) in a 1:10 ratio and 1 mL of homogenate was transferred to 11 tubes. These homogenates were seeded with serial dilutions (1.0 × 108, 1.0 × 107, 1.0 × 106, 1.0 × 105, 1.0 × 104, 1.0 × 103, 500, 250, 125, 62, 31 CFU/mL) of a mixture of pure cultures of three bacterial species. DNA was extracted from 200 µL of each dilution as described above. Extracted DNAs obtained from the dilutions with the same dilution factor of each bacterial suspension were mixed together and used as template DNA.
The m-PCR method was developed to detect three bacteria, Y. ruckeri, A. salmonicida subsp. salmonicida and F. psychrophilum simultaneously using different combinations of the five specific primer pairs and the specificity of this method was evaluated in three different m-PCR trials as follows.
In Trial 1, the expected sizes of the PCR products were obtained using the primers mentioned above. However, of all the A. salmonicida subsp. salmonicida strains, strains No. 19 and No. 22 could not be detected using PAAS1/2 primers and No. 19 was detected to give a non-specific band. The other strains used in this study did not present any non-specific or false specific bands. A. salmonicida subsp. salmonicida strain No. 19 and No. 22 could also not be detected using PAAS1/2 primers and A. hydrophila strain No. 26 was detected to give a false specific band with these primers in Trial 2. The other strains gave the expected size of bands and they did not show any non-specific or false specific bands. The molecular weight of the products obtained from amplification using ruck1/2 and PAAS1/2 for Yersinia ruckeri and A. salmonicida subsp. salmonicida, respectively, were detected to be close each other.
In Trial 3, YER8/10, Fer3/4 and FP1/3 primers were used for Y. ruckeri, A. salmonicida subsp. salmonicida and F. psychrophilum, respectively. The size of the PCR products obtained for F. psychrophilum, Y. ruckeri and A. salmonicida were 971 bp, 575 bp and 422 bp, respectively (Fig. 1). A. salmonicida subsp. salmonicida strains which could not be detected in the two other trials were successfully detected in this trial. None of the strains used in this study showed false negative or non-specific bands. Fer3/4 primers used for Aeromonas salmonicida subsp. salmonicida strains were determined to have more specificity than PAAS1/2 primers.
To optimize the m-PCR method, the amplification protocol was reorganized with the primer sets of Trial 3. In accordance with the new amplification protocol, PCR conditions consisted of an initial denaturation at 94℃ for 3 min followed by 30 cycles of amplification in which denaturation, annealing and extension temperatures were 94℃ for 45 sec, 60℃ for 45 sec and 72℃ for 45 sec, respectively. A final extension step was at 72℃ for 3 min. At the end of this protocol, all strains used in this study were detected to give amplicons with their specific primer sets and not to give false specific or non-specific bands. Y. ruckeri, A. salmonicida subsp. salmonicida and F. psychrophilum could be detected in a shorter time with this new protocol.
To optimize the oligonucleotide primers, we used 1, 0.8, 0.5, 0.3, 0.1 and 0.05 µM concentrations of each primer. The expected bands were observed even at the minimum 0.05 µM concentration of primers. However, we detected that A. hydrophila strain No. 28 gave a 575 bp product specific to Y. ruckeri at <1 µM concentration of primer and also gave a 452 bp product specific to A. salmonicida subsp. salmonicida at <0.5 µM concentration of primer. Thus, the optimal primer concentration was considered to be 1 µM.
In the optimization study for Taq polymerase, we used 1 µM of each oligonucleotide primer and 0.35, 0.25, 0.15, 0.1, 0.05 and 0.02 U of Taq polymerase. We observed that Y. ruckeri and A. salmonicida subsp. salmonicida could be detected with 0.05 U Taq polymerase, while F. psychrophilum could be detected with 0.15 U Taq polymerase. Furthermore the minimum concentration of Taq polymerase used in the simultaneous detection of these bacteria was found to be 0.15 U. The intensity of PCR bands observed was determined to be better at 0.25 U concentration of Taq polymerase.
In the optimization of MgCl2, we used 1 µM of each oligonucleotide primer, 0.25 U of Taq polymerase and 2, 1.5, 1, 0.7, 0.4 and 0.2 mM of MgCl2. The minimum concentrations of MgCl2 for simultaneous detection of these three bacteria were found to be 1 mM for Y. ruckeri and A. salmonicida subsp. salmonicida, and 1.5 mM for F. psychrophilum. Also, 2 mM of MgCl2 was determined to produce a more intense band for each target DNA. Thus, 2 mM concentration of MgCl2 was selected for optimal m-PCR condition.
The best results were obtained with an initial denaturation of 25 µL PCR mixture containing DEPC-treated water, 1× PCR buffer, 2 mM MgCl2, 0.2 mM of each dNTP, 0.25 U Taq polymerase, 1 µM of each primer and 3 µL of template DNA at 94℃ for 3 min, followed by 30 cycles of amplification in which denaturation, annealing and extension temperatures were 94℃ for 45 sec, 60℃ for 45 sec and 72℃ for 45 sec, respectively, and a final extension step at 72℃ for 3 min (Fig. 1).
The detection limits of DNA were found to be 35 pg of DNA for Y. ruckeri and F. psychrophilum, 70 pg of DNA for A. salmonicida subsp. salmonicida (Fig. 2).
The bacterial enumeration was performed and the logarithmic phase stock cultures prepared for each bacterium were adjusted to 1.0 × 108 CFU/mL. The genomic DNA extracted from the serial dilutions (1.0 × 107, 1.0 × 106, 1.0 × 105, 1.0 × 104, 1.0 × 103, 500, 250, 125, 62, 31, 15, 7 CFU/mL) of each suspension were used in the m-PCR assay developed in this study and the assay could detect as few as 15 CFU of Y. ruckeri and F. psychrophilum and 30 CFU of A. salmonicida subsp. salmonicida in pure cultures (Fig. 3). Also, the detection limit in the presence of tissue debris was found to be 125 CFU for Y. ruckeri and F. psychrophilum, and 250 CFU for A. salmonicida subsp. salmonicida (Fig. 4).
Aquaculture is currently one of the fastest growing food production system in the world. Under intensive aquaculture conditions the risk of stress increases and significant proportions of stock can become infected. Especially, healthy looking fish without any lesions or clinical signs of disease can carry some pathogens and create a serious risk for spread of contagious diseases in fish populations. The detection of pathogen carrier fish is thus essential for effective fish disease control. To detect carrier fish, a sufficiently sensitive, specific and cost-effective system is required. Such a system could be useful in monitoring fish populations [1,3].
Of all the primers evaluated for their specificity in Trials 1~3, PAAS1/2 primers were shown to be less capable of detecting some A. salmonicida subsp. salmonicida strains than the Fer3/4 primers used in Trial 3. A nonspecific band also occurred with PAAS1/2. Therefore, we decided to discard these primer pairs. Ruck1/2 primers used for Y. ruckeri were also discarded because the molecular size of amplification products obtained using ruck1/2 and PAAS1/2 were too close not to allow discrimination of Y. ruckeri and A. salmonicida subsp. salmonicida. However, some researchers found no false positive results with PAAS1/2 in their PCR assays and these primers were specific for their target, A. salmonicida [1,6]. Taking into consideration these previous results, the use of ruck1/2 and PAAS1/2 primers in the combinations of primer sets for Y. ruckeri and A. salmonicida subsp. salmonicida, respectively, was inevitably inappropriate for our m-PCR assay. At the end of Trial 3 which was intended to determine the specificity of YER8/10, Fer3/4 and FP1/3 primers, neither nonspecific nor false specific amplification product was observed. Therefore, these primers were proved to be specific under the conditions designed for this assay. DNA of neither closely related bacteria nor other unrelated bacteria used in this study were amplified in this multiplex assay. Thus, we demonstrated that this assay has a high specificity. Beaz-Hidalgo et al. [5] have also reported that Fer3/4 primers have high specificity to detect A. salmonicida. The YER 8/10 primers used in this study for amplification of Y. ruckeri in m-PCR system have also been reported to be adequate for specific detection of Y. ruckeri [8].
In the m-PCR assay developed in this study, the detection limit was 15 CFU for Y. ruckeri and F. psychrophilum and 30 CFU for A. salmonicida subsp. salmonicida in pure cultures. The detection limits in the same protocol were 35 pg for Y. ruckeri and F. psychrophilum, and 70 pg for A. salmonicida subsp. salmonicida. The detection limit in the presence of tissue debris was found to be 125 CFU for Y. ruckeri and F. psychrophilum, and 250 CFU for A. salmonicida subsp. salmonicida. Byers et al. [6] have determined that the PAAS PCR had an in vitro sensitivity of 93%, and their 0.6% false negative rate might have been related to the primer target site which was believed to be present in approximately 90% of A. salmonicida isolates. In our study, of all 9 strains tested, 2 strains were unidentified in m-PCR assay using PAAS1/2, so 77.7% of the A. salmonicida strains tested in this study were identified correctly. Although Byers et al. [6] have stated the detection limit of PAAS PCR to be in the range 2 pg to 0.2 pg per PCR, we were unable to obtain as low as their values. Altinok et al. [1] have reported that the detection limits of A. salmonicida and Y. ruckeri which were 2 of 5 target bacteria for detection in a m-PCR assay were 1 CFU for A. salmonicida and Y. ruckeri in pure culture, and 5 and 4 CFU for A. salmonicida and Y. ruckeri, respectively, in the presence of fish tissue. Gibello et al. [8] have reported the detection limit of a single PCR procedure for Y. ruckeri to be 60~65 cells per PCR mixture in culture, and 2 × 104 CFU/g in tissues. The detection limits in our study for Y. ruckeri were found to be lower.
There is growing interest in the diagnosis of important fish diseases in culture fisheries. The progress in molecular techniques aids the rapid identification of causative agents of these diseases. In conclusion, the m-PCR assay which developed in this study for the simultaneous detection of three major pathogens, Y. ruckeri, A. salmonicida subsp. salmonicida and F. psychrophilum, was proved to be highly specific for detecting these three bacteria. We submit that rapid and reliable identification of these bacteria can be achieved by this m-PCR assay.
Figures and Tables
Fig. 1
Simultaneous identification of Flavobacterium (F.) psychrophilum (971 bp), Yersinia (Y.) ruckeri (575 bp), and Aeromonas (A.) salmonicida subsp. salmonicida (422 bp) by m-PCR3. Lane M: Molecular size marker (100~1,000 bp), Lane 1: m-PCR with the three pathogens together, Lane 2: A. salmonicida subsp. salmonicida alone, Lane 3: Y. ruckeri alone, Lane 4: F. psychrophilum alone, Lane 5: F. psychrophilum and Y. ruckeri, Lane 6: F. psychrophilum and A. salmonicida subsp. salmonicida, Lane 7: Y. ruckeri and A. salmonicida subsp. salmonicida.
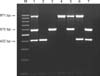
Fig. 2
Determination of detection limit of m-PCR assay. Lane M: Molecular size marker (100~1,000 bp), Lane a-h: the serial dilutions of chromosomal DNA extracted from reference F. psychrophilum, Y. ruckeri and A. salmonicida subsp. salmonicida strains, from left to right, 1,055, 700, 350, 210, 140, 70, 35 and 17 pg DNA, respectively.

Fig. 3
Detection limit of m-PCR assay in pure cultures of bacteria. Lane M: Molecular size marker (100~1,000 bp), Lane 1: 1.0 × 103, Lane 2: 500, Lane 3: 30, Lane 4: 15 CFU of F. psychrophilum, Y. ruckeri and A. salmonicida subsp. salmonicida, respectively.

Fig. 4
The detection limit of m-PCR assay in the presence of tissue debris. Lane M: Molecular size marker (100~1,000 bp), Lane 1: 1.0 × 105, Lane 2: 1.0 × 104, Lane 3: 1.0 × 103, Lane 4: 250 Lane 5: 125 CFU of F. psychrophilum, Y. ruckeri and A. salmonicida subsp. salmonicida, respectively.
Acknowledgments
This study (Grant No. 107O208) was supported by the Scientific and Technological Research Council of Turkey (TUBITAK). We thank to Dr. Jessica Boyd (NRC Institute for Marine Biosciences, Canada) and Prof. Dr. Hasmet Cagirgan (Ege University, Faculty of Fisheries, Turkey) for providing certain strains of A. salmonicida subsp. salmonicida and Y. ruckeri.
References
1. Altinok I, Capkin E, Kayis S. Development of multiplex PCR assay for simultaneous detection of five bacterial fish pathogens. Vet Microbiol. 2008. 131:332–338.

2. Altinok I, Grizzle JM, Liu Z. Detection of Yersinia ruckeri in rainbow trout blood by use of the polymerase chain reaction. Dis Aquat Organ. 2001. 44:29–34.
3. Altinok I, Kurt I. Molecular diagnosis of fish diseases: a review. Turk J Fish Aquat Sci. 2003. 3:131–138.
4. Baliarda A, Faure D, Urdaci MC. Development and application of a nested PCR to monitor brood stock salmonid ovarian fluid and spleen for detection of the fish pathogen Flavobacterium psychrophilum. J Appl Microbiol. 2002. 92:510–516.

5. Beaz-Hidalgo R, Magi GE, Balboa S, Barja JL, Romalde JL. Development of a PCR protocol for the detection of Aeromonas salmonicida in fish by amplification of the fstA (ferric siderophore receptor) gene. Vet Microbiol. 2008. 128:386–394.

6. Byers HK, Gudkovs N, Crane MSJ. PCR-based assays for the fish pathogen Aeromonas salmonicida. I. Evaluation of three PCR primer sets for detection and identification. Dis Aquat Organ. 2002. 49:129–138.

7. del Cerro A, Mendoza MC, Guijarro JA. Usefulness of a TaqMan-based polymerase chain reaction assay for the detection of the fish pathogen Flavobacterium psychrophilum. J Appl Microbiol. 2002. 93:149–156.

8. Gibello A, Blanco MM, Moreno MA, Cutuli MT, Domenech A, Domínguez L, Fernández-Garayzábal JF. Development of a PCR assay for detection of Yersinia ruckeri in tissues of inoculated and naturally infected trout. Appl Environ Microbiol. 1999. 65:346–350.

9. Gustafson CE, Thomas CJ, Trust TJ. Detection of Aeromonas salmonicida from fish by using polymerase chain reaction amplification of the virulence surface array protein gene. Appl Environ Microbiol. 1992. 58:3816–3825.

10. Kodama H, Honda A, Mikami T, Izawa H. Detection of fish antibody against protein antigen of Aeromonas salmonicida by enzyme-linked immunosorbent assay using biotin-avidin system. Res Vet Sci. 1987. 43:78–84.

11. LeJeune JT, Rurangirwa FR. Polymerase chain reaction for definitive identification of Yersinia ruckeri. J Vet Diagn Invest. 2000. 12:558–561.

12. Loghothetis PN, Austin B. Antibody responses of rainbow trout (Oncorhynchus mykiss, Walbaum) to live Aeromonas hydrophila as assessed by various antigen preparations. Fish Shellfish Immunol. 1996. 6:455–464.

13. Mata AI, Gibello A, Casamayor A, Blanco MM, Domínguez L, Fernández-Garayzábal JF. Multiplex PCR assay for detection of bacterial pathogens associated with warm-water Streptococcosis in fish. Appl Environ Microbiol. 2004. 70:3183–3187.

14. Michel C, Antonio D, Hedrick RP. Production of viable cultures of Flavobacterium psychrophilum: approach and control. Res Microbiol. 1999. 150:351–358.

15. O'Brien D, Mooney J, Ryan D, Powell E, Hiney M, Smith PR, Powell R. Detection of Aeromonas salmonicida, causal agent of furunculosis in salmonid fish, from the tank effluent of hatchery-reared Atlantic salmon smolts. Appl Environ Microbiol. 1994. 60:3874–3877.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download