Abstract
In August 2008, forty dogs out of 400 developed oral warts in a breeding farm in Korea. Canine oral papilloma infection is a common disease in dogs. However, there has been no report of an outbreak of canine oral papillomavirus (COPV) in a group of dogs or in dog breeding farms in Korea, and the genetic analysis of COPV in Korea has yet to be performed. This study diagnosed canine oral papilloma from the oral samples of these dogs based on histopathological examination and immunohistochemistry. Polymerase chain reaction was applied to amplify the corresponding products using pre-existing primer sets for COPV and a universal human papillomavirus targeting L1 gene. Further genetic analysis of the major viral capsid gene L1 confirms the sequences of Korean COPV, which shows a close relationship to previously reported COPV. This study describes the histopathological and immunohistochemical characteristics of canine oral papilloma in a group of breeding dogs in Korea and discloses the complete L1 gene sequences of Korean COPV.
Papillomaviruses cause warts in various animals including dogs [3], and human papillomaviruses (HPVs) are important because of their causative role in cervical cancer [17]. Canine oral papilloma mainly affects young dogs, approximately 1 year old in age, and there is no difference in prevalence between sex and breed [6]. This tumor can be diagnosed easily with gross morphological and histopatholgical characteristics. The tumors grow exophilic and have a "cauliflower-like surface" [6]. In histopathology, fibrovascular core is prominent, and some large cells in the stratum granulosum can display amphophilic intranuclear inclusion body instead of nuleoli [6].
The papillomavirus genome consists of early gene and late gene regions. The early viral proteins E1 to E7 play roles in replication of the viral genome or control cell cycle to enhance viral DNA replication [5,8,17]. L1 and L2 which form the viral capsid and package viral DNA are the late proteins [16]. The L1 and L2 genes which translate these late proteins comprise a majority of total viral DNA. Moreover, the L1 gene of HPV was known to a high degree of nucleotide sequence identity [4]. Although only limited sequence information is available for the complete genome or the L1 gene of canine oral papillomavirus (COPV), it could be predicted that L1 gene also shows a characteristically high degree of nucleotide sequence identity like HPV because the L1 gene of other papillomaviruses was the most conserved gene among papillomavirus genomes, and phylogenetic analyses of the L1 gene have been broadly used for the classification of papillomaviruses including canine papillomaviruses [2,15].
In August 2008, forty dogs out of 400 in a breeding farm showed oral warts. The neoplasmas which have "cauliflower-like surface" [6], were white to gray color and the diameter of the mass was about 1~2 cm. The tumor regression took 4~8 weeks, and no oral carcinomas developed. The papilloma type tumors were reported as low percentage in Korea. However, there has been no report of an outbreak in a group of dogs or dog breeding farms in Korea, and genetic analysis of Korean COPV has yet to be performed. The aim of this study was to confirm the massive outbreak of canine papilloma in a breeding farm in Korea using histopathological and immunohistochemical analyses and to describe the complete sequence of the L1 gene of Korean COPV.
This study used oral neoplastic tissues of 7 representive patients who had canine papilloma. The 40 patients were all 4 months old and all Korean mongrel dogs. At approximately 3.5 months of age, they began to show oral warts which slowly increased in number and size. Biopsies were performed on 7 representative dogs around 4 months of age. The biopsy specimens were collected from the tongue, buccal skin, and nose of seven dogs. The tissues were divided into 2 pieces, and each piece was fixed in 10% neutral buffered formalin or frozen at -70℃.
The formalin fixed oral specimens were embedded in paraffin, and 4 µm sections cut from each paraffin block. Sections were stained with hematoxylin and eosin and diagnosed.
For immunohistochemical analysis, polyclonal rabbit papillomavirus antibody (Dako North America, USA) was used. The slides were deparaffinized, rehydrated, and treated with a 3% hydrogen peroxide (H2O2) solution for 20 min at room temperature. After washing in phosphate-buffered saline (PBS) three times, the antigens were retrieved by boiling the sections in citric acid buffer (pH 6) for 10 min in the microwave oven (high power). Sections were incubated with primary antibodies for 2 h. The secondary polymer of Envision system-HRP (LSAB kit; Dako North America, USA) was applied to each slide for 30 min, and the slides were washed 4 times with PBS. Next, the slides were incubated with substrates until desired staining intensity developed. The color reaction was stopped by washing in distilled water twice, and counterstained with Harris hematoxylin.
Polymerase chain reaction (PCR) was applied to frozen tissues to compare with the genome of known papillomaviruses. Three different primers used in this study were selected from the primers information by others [4,9,14]. Primer sets 1 and 2 were specific for the diagnosis COPV [14] or canine cutaneous papillomavirus [9], respectively. Primers set 3 consisted of degenerate primers able to amplify a broad range of human papillomavirus L1 genes [4].
Freshly collected oral neoplastic tissues of canine papilloma patients were promptly frozen at -70℃ and stored until processing. To prepare DNA, approximately 100 mg of tissue samples were thawed, minced, and homogenized in microtubes. DNAzol Reagent (Invitrogen, USA) was used to extract DNA according of the manufacturer's instructions. The PCR reaction was carried out with the Maxime PCR premix kit (Intron, Korea), and each amplification followed the conditions as previously described [4,9,14]. The PCR products were analyzed by electrophoresis in a 2.0% agarose gel stained with ethidium bromide and observed under UV transillumination. PCR products were sequenced with ABI PRISM BigDye Terminator v3.0 Cycle sequencing kit (Applied Biosystems, USA).
In addition, for partial sequence comparison with other COPV or papillomaviruses, another set of primers targeting whole gene of a major structural gene L1 were designed based on the available COPV sequence information (GenBank Accession numbers D55633, D26115, L22695 and NC_001919). The forward primer was 5'-TGACATTTTGGACTCCTCTGC, and the reverse primer was 5'-TGCCGGTCAGTCAGAAACA. The PCR reaction was carried out with HotStarTaq Plus DNA polymerase (Qiagen, Germany), and the sequencing method was the same above.
The papillomas affected the buccal mucosa, lips, tongue, and nose (Fig. 1). The neoplasm formed papillary or verruciform protrusions. The "cauliflower-like surface" [6] masses were white to gray color with an approximate diameter of 1~2 cm, and the masses in the oral cavity were bigger than those of on the nose or lips. The firm nodules grew outward and were peduncular, which is a characteristic appearance of papilloma.
The epithelial proliferation was prominent, and the thickness of the epidermis was increased. Thin papillary projections and numbers of keratohyalin-like granules were observed (Fig. 2). The nuclei of the keratinocytes in the stratum granulosum or spinosum were large, and some cells in the stratum granulosum losing nucleoli display amphophlic intranuclear inclusion bodies (Fig. 3). The keratohyalin granules of up to nuclear size were observed in the keratinocytes or stratum corneum.
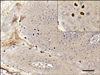
Some nuclei of keratinocytes in the proliferated epidermis had a positive reaction with bovine papillomavirus antibody. The nuclei with positive reactions were prominent just beneath the corneal layer, but not all keratinocytes nuclei of the affected area were positive for papillomavirus antibody (Fig. 4). The stratum corneum, which consists of keratin, also had strong positive immunolabeling with the antibody in some parts of the affected area.
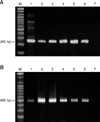
In PCR amplification, primer set 1 amplified 261 base pairs (bps) for the L1 gene region of COPV (Fig. 5A) and primer set 3, which amplifies 478-483 bps nucleotides of broad range of human HPVs, also produced amplicons from the DNA of the tested samples. Amplicons were detected by gel electrophoresis, and 6 samples yielded similar sizes of bands (Fig. 5B). Based on sequencing of PCR products, the size of actual amplicons was confirmed as 493 bps, which was the same length and sequence as previously known sequences of COPV (GenBank Accession Nos. D26115, D55633, L22695 and NC_001919) [3,7]. PCR using primers targeting whole gene of a major structural gene L1 amplified approximately 1.6 kb and the comparison of nucleotide sequences of the entire L1 gene completely matched previously reported sequences of COPV (data not shown). The L1 gene sequence of Korean COPV (KCOPV1) was deposited in GenBank (Accession No: FJ479789). On the other hand, the 194 bp band was not detectable after amplification with primer set 2 for canine cutaneous papillomavirus (data not shown).
The results of this study confirm that the massive outbreak of oral warts from 40 dogs in Korea in 2008 was due to COPV. The histological changes and the result of IHC were almost the same with previously known disease information [6,10,11] such as gross morphology of the mass, epithelial proliferation, ballooning degeneration of keratinocytes with intranuclear inclusion bodies, and reactivity with bovine papillomaviral antibody.
Seven tested samples showed histopathological changes of COPV infection, and all tested samples (7/7) were immunoreactive with bovine papillomavirus antibody with immunohistochemically labeled nuclei. PCR amplification with primer sets 1 and 3 both yielded the expected fragments of approximately 260 and 480 bps in 6 cases, but the PCR results with primer sets 1 and 3 were both negative in case of No.7. The negative result may be due to the small amount of viral DNA in the sample. PCR is very sensitive tool for diagnosis (85.7%), but IHC results appeared to be more sensitive in this study (100%). The antibody used in this study was originally prepared for detecting viral structural antigens (viral capsid L1 protein antigens) of bovine papillomavirus (BPV). Also, a rabbit polyclonal antibody for BPV is known to cross-react with canine cutaneous papillomavirus [1,9,12]. In this study, we were able to detect viral antigens with BPV antibody in tissues affected by canine oral papilloma. This implies that BPV antibody cross-reacts with both cutaneous and oral canine papillomaviruses, possibly due to common epitope(s) in the L1 capsid protein, even though a previous study described that the nucleotide sequences of L1 gene of canine cutaneous papillomavirus showed less than 57% sequence similarity to COPV [13]. The immune-reactive nuclei of keratinoytes tend to be localized just beneath the stratum corneum, and the viral capsid L1 protein antigen was also detected in the keratin layer of the affected area.
In this study, three kinds of pre-existing primers for papillomavirus were used for the primary diagnosis. Both primer sets for COPV (primer set 1) and universal HPV (primer set 3) targeting the L1 gene amplified their corresponding bands. Even though the universal degenerate primer sets were originally designed to detect a broad range of cutaneous HPVs and the COPV showed low sequence identity (≤60%) of the L1 gene with most known HPVs (data not shown), L1 is the most highly conserved gene within all other known papillomaviruses [2]. Therefore, both primer sets can be used as good diagnostic tools for canine oral and possibly cutaneous papillomaviruses. However, the primer set for cutaneous canine papillomavirus seemed to be very specific for cutaneous type of papilloma since these primers did not generate any visible bands. The failure to amplify products was probably predictable because the samples that were used for this study were collected from the frank regions of oral warts and the low sequence similarity of the canine cutaneous papillomavirus L1 gene with COPV [13].
COPVs isolated from papilloma tissues can be easily transmitted from dog to dog in the laboratory [7]. A previous study also described that papillomatosis occurring in experimental dog colonies affected up to 25% of animals [11]. In this study, only 40 (10%) of 400 dogs showed clinical signs of oral warts by natural infection, although there were transmissible opportunities for others to infect COPV.
In summary, the histopathological and genetic analysis confirmed the outbreak of canine oral warts and the causative agent as COPV. This study describes the outbreak of canine oral papilloma infection in a group of Korean breeding dogs and discloses the complete L1 gene sequence information of Korean COPV. Further studies need to be performed to investigate the prevalence of COPV and/or canine cutaneous papillomavirus in Korea by using these diagnostic techniques.
Figures and Tables
 | Fig. 1Gross appearance of the oral warts. The papillomas with verruciform protrusions affected the buccal mucosa, lips, and nose. |
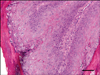 | Fig. 2Papillomatous proliferation of epidermal cells and keratohyalin-like granules are present in the stratum corneum and cytoplasms of keratinocytes. H&E stain. Scale bar = 75 µm. |
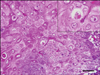 | Fig. 3Large nuclei of the keratinocytes and amphophlic intranuclear inclusion bodies are observed in the keratinocytes. H&E stain. Scale bar = 37.5 µm. |
References
1. Campbell KL, Sundberg JP, Goldschmidt MH, Knupp C, Reichmann ME. Cutaneous inverted papillomas in dogs. Vet Pathol. 1988. 25:67–71.

2. de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004. 324:17–27.

3. Delius H, Van Ranst MA, Jenson AB, zur Hausen H, Sundberg JP. Canine oral papillomavirus genomic sequence: a unique 1.5-kb intervening sequence between the E2 and L2 open reading frames. Virology. 1994. 204:447–452.

4. Forslund O, Antonsson A, Nordin P, Stenquist B, Hansson BG. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J Gen Virol. 1999. 80:2437–2443.

5. Goldstein DJ, Finbow ME, Andresson T, McLean P, Smith K, Bubb V, Schlegel R. Bovine papillomavirus E5 oncoprotein binds to the 16K component of vacuolar H+-ATPases. Nature. 1991. 352:347–349.

6. Head KW, Else RW, Dubielzig RR. Meuten DJ, editor. Tumors of the alimentary tract. Tumors in Domestic Animals. 2002. 4th ed. Ames: Iowa State University Press;422–423.

7. Isegawa N, Nakano K, Ohta M, Shirasawa H, Tokita H, Simizu B. Cloning and sequencing of the L1 gene of canine oral papillomavirus. Gene. 1994. 146:261–265.

8. Masterson PJ, Stanley MA, Lewis AP, Romanos MA. AC-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase alpha-primase p68 subunit. J Virol. 1998. 72:7407–7419.

9. Narama I, Kobayashi Y, Yamagami T, Ozaki K, Ueda Y. Pigmented cutaneous papillomatosis (pigmented epidermal nevus) in three pug dogs; histopathology, electron microscopy and analysis of viral DNA by the polymerase chain reaction. J Comp Pathol. 2005. 132:132–138.

10. Nicholls PK, Klaunberg BA, Moore RA, Santos EB, Parry NR, Gough GW, Stanley MA. Naturally occurring, nonregressing canine oral papillomavirus infection: host immunity, virus characterization, and experimental infection. Virology. 1999. 265:365–374.

11. Nicholls PK, Stanley MA. Canine papillomavirus - A centenary review. J Comp Pathol. 1999. 120:219–233.
12. Shimada A, Shinya K, Awakura T, Narama I, Maeda H, Umemura T. Cutaneous papillomatosis associated with papillomavirus infection in a dog. J Comp Pathol. 1993. 108:103–107.

13. Tanabe C, Kano R, Nagata M, Nakamura Y, Watanabe S, Hasegawa A. Molecular characteristics of cutaneous papillomavirus from the canine pigmented epidermal nevus. J Vet Med Sci. 2000. 62:1189–1192.

14. Teifke JP, Löhr CV, Shirasawa H. Detection of canine oral papillomavirus-DNA in canine oral squamous cell carcinomas and p53 overexpressing skin papillomas of the dog using the polymerase chain reaction and non-radioactive in situ hybridization. Vet Microbiol. 1998. 60:119–130.

15. Yuan H, Ghim S, Newsome J, Apolinario T, Olcese V, Martin M, Delius H, Felsburg P, Jenson B, Schlegel R. An epidermotropic canine papillomavirus with malignant potential contains an E5 gene and establishes a unique genus. Virology. 2007. 359:28–36.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download