Abstract
Despite global efforts to control porcine reproductive and respiratory syndrome virus (PRRSV) infection, the virus continues to cause economic problems in the swine industry worldwide. In this study, we attempted to generate and characterize a panel of stable BHK cell lines that constitutively express the nucleocapsid (N) protein of type 1 or type 2 PRRSV. The established BHK cell lines were found to react well with N-specific antibodies as well as the hyperimmune serum of pigs raised against each genotype of PRRSV. Taken together, the data implicate a potential usefulness for the newly generated stable cell lines as a diagnostic reagent for PRRSV serology.
Porcine reproductive and respiratory syndrome (PRRS) emerged almost simultaneously in North America and Europe. This disease has since plagued nearly all pig-producing countries with a massive economic impact on the swine industry worldwide [7]. The causative agent, PRRS virus (PRRSV), is a small enveloped, single-stranded, positive-sense RNA virus belonging to the family Arteriviridae in the order Nidovirales [9]. PRRSV is divided into two major genotypes, the European (EU; type 1) genotype and the North American (NA; type 2) genotype. These two genotypes were formerly located on different continents [5] but intermingled existence of the two genotypes has now been identified in many parts of the world, including Korea [4,8]. This situation leads to potentially problematic consequences for PRRSV diagnostics and management, since current serological tools are usually incapable of identifying whether a pig is infected with a single genotype or co-infected with two genotypes.
The nucleocapsid (N) protein of PRRSV encoded in ORF7 is a small basic multifunctional protein with a molecular weight of 15-kDa [9]. The PRRSV N protein is the most abundant protein in infected cells, constituting approximately 40% of the protein content of the virion, and is known to be highly immunogenic in the natural host [3]. Furthermore, the N protein is encoded by a region of the viral genome that is relatively well conserved between the two genotypes [6]. Thus, these properties enable the N gene to be a proper candidate for the detection of antibodies against PRRSV. In the present study, we aimed to generate a panel of stable BHK cell lines constitutively expressing N proteins from EU and NA genotypes of PRRSV and subsequently to assess their potential use as a diagnostic reagent in immunofluorescence assay (IFA) and immunoperoxidase monolayer assay (IPMA) techniques.
The viral RNA was extracted from each virus stock of Korean type 1 and 2 PRRSV strains, KNU-07 and PL97-1 [1,4], by using the Viral RNA Mini Kit (Qiagen, Germany). RT-PCR was performed to amplify the full-length ORF7 gene from EU (KNU-07) and NA (PL97-1) genotypes with the following corresponding primer pairs: KNU07-ORF7-F (5'-GCCGGGATCCACCATGGCCGGTAGAAAC-3'), KNU07-ORF7-R (5'-GCCGGGATCCATTTGCATCCTGACTGG-3'), PL97-ORF7-F (5'-GCCGGGATCCACCATGCCAAATAACAACGGC-3') and PL97-ORF7-R (5'-GCCGGGATCCTGCTGAGGGTGATGC-3'), where underlines indicate the BamHI restriction enzyme sequence. Each PCR amplicon was initially inserted individually into a pBudCE4.1 Vector (Invitrogen, USA) that contains six repetitive histidine codons. Individual His-tagged PRRSV ORF7 cDNA fragments were then subcloned independently into a pFB-Neo Retroviral Vector (Stratagene, USA) using SalI and EcoRI restriction sites.
The retrovirus gene transfer system (Stratagene, USA) was applied to generate BHK cell lines constitutively expressing the recombinant ORF7 gene [2]. The selected cell clones (BHK-EU-ORF7 and BHK-NA-ORF7) in the presence of 800 µg/ml G418 (Invitrogen, USA) were initially subjected to PCR to identify the PRRSV ORF7 gene integration, followed by RT-PCR and nucleotide sequencing to determine N gene expression at the mRNA level. The full-length N genes of about 400 bp were identifiable from individual BHK-EU-ORF7 and BHK-NA-ORF7 cell clones (data not shown). Next, N protein expression at the protein level was tested by western blot analysis [2] and robust levels of approximately 15-kDa N were detected in all selected BHK cell clones (Figs. 1A and B, upper panels). Each BHK-EU-ORF7 or BHK-NA-ORF7 cell clone that expressed the highest level of N protein was chosen for subsequent studies.
The BHK-EU-ORF7 and BHK-NA-ORF7 cell lines were further examined for the subcellular expression of N gene by IFA [2]. As expected, the specific cytoplasmic and nucleolus staining was clearly evident when the cells were reacted with the anti-N monoclonal antibody (MAb) SDOW17, confirming the constant high level expression of the N protein. In addition, the overall growth kinetics of each N gene-expressing cell line was found to be similar to that of the parental BHK-21 cells, indicating that the N expression has no effect on cell growth (data not shown). This result further suggests their potential use as a suitable source for cell-based diagnostic tools.
In order to assess whether the PRRSV N gene-expressing stable BHK cell lines are potentially applicable for PRRSV serology, we first conducted IFA employing genotype-specific MAbs raised against the N protein (anti-EU, anti-NA, anti-EU + NA; GenoBiotech, Korea). BHK-EU-ORF7 cells expressing the N gene of the EU genotype were shown to react well with the EU genotype-specific MAb (Fig. 2A); conversely, they inefficiently recognized the NA genotype-specific MAb, indicating the presence of a minimal cross-reaction (Fig. 2B). For BHK-NA-ORF7 cells expressing the N gene of the NA genotype, the results were similar to those obtained from BHK-EU-ORF7 cells, with the specific fluorescence of the cells incubated with the NA genotype-specific MAb (Fig. 2E). In addition, both BHK-EU-ORF7 and BHK-NA-ORF7 cells were well reactive with the N-specific MAb (α-EU + NA) commonly responding to both PRRSV genotypes (Figs. 2C and F). Taken together, the data suggest the potential capability of these stable cells lines for the detection and discrimination of the genotypes.
Our diagnostic approaches were further extended to IPMA in stable BHK cell lines expressing each of the N genotypes. At 2 days post-seeding, BHK-EU-ORF7 or BHK-NA-ORF7 cells co-cultivated with normal BHK cells at 1 : 1 ratio in 96-well tissue culture plates were fixed with cold methanol and subjected to immunoperoxidase staining. Although some cross-reactivity occurred in BHK-EU-ORF7 cells (Fig. 3B) that was consistent with the IFA result (Fig. 2B), both stable cell lines reacted specifically with N-specific antibodies by IPMA (Figs. 3A-F). These data indicated that IPMA using the N gene-expressing BHK cell lines could be an alternative antibody detection method. To further examine whether IPMA is valuable for detecting serum antibody response from animals infected with various genetically different field strains, the hyperimmune pig serum raised against either type 1 (LV) or type 2 (VR-2332) PRRSV was used for this antibody detection test. As shown in Fig. 3, both BHK-EU-ORF7 and BHK-NA-ORF7 cells were well reactive with PRRSV-specific hyperimmune sera (G, H, J, and K). However, we found cross-reactions of two different genotype-specific pig sera in both cell lines (Figs. 3H and J). These consequences may be non-specific or due to unknown porcine serum components and, accordingly, the tests should be optimized for field serum samples. In contrast, no stained cells were observed in N gene-expressing cells incubated with the pre-immune negative serum (Figs. 3I and L).
In conclusion, we report here the development of stable BHK cell lines constitutively expressing the genotype-specific PRRSV N protein that reacted well with various N-specific antibodies as well as anti-PRRSV hyperimmune pig sera. Given these observations, we suggest that these N gene-expressing cells would allow the simultaneous detection and differentiation of antibodies against the EU and NA genotypes of PRRSV. IFA and IPMA methods using N-expressing cells are a low cost alternative to a more expensive commercial IDEXX ELISA kit and consequently are considered to be potential tools for routine serological tests and epidemiological surveys. Additionally, future comparative analysis with the commercial ELISA method using field pig serum samples is definitely needed to investigate sensitivity and specificity of the cell line-based IFA and IPMA. Along with being a valuable diagnostic reagent for detecting PRRSV antibodies in pigs, established cell lines expressing the genotype-specific N protein may serve as potential sources of diagnostic antigens in ELISA or as a research tool for N-specific antibody production or vaccination studies.
Figures and Tables
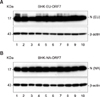 | Fig. 1Constitutive expression of the genotype-specific nucleocapsid (N) protein in BHK-EU-ORF7 (A) and BHK-NA-ORF7 (B) cells. Expression levels of the N protein in individual stable cell clones were determined by western blot analysis. Each lane represents individual neomycin-resistant BHK cell clones. |
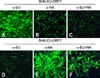 | Fig. 2Immunofluorescence assay using the genotype-specific monoclonal antibodies. BHK-EU-ORF7 and BHK-NA-ORF7 cells were incubated individually with each genotype-specific antibody. A and D α-EU: EU genotype-specific monoclonal antibody (MAb), B and E α-NA: NA genotype-specific MAb, C and F α-EU + NA: genotype-common MAb. ×40. |
 | Fig. 3Immunoperoxidase monolayer assay. Cells were incubated with the genotype-specific MAbs or PRRSV-specific hyperimmune sera followed by staining using Vectorstain ABC peroxidase and DAB substrate kits. G and J α-LV: anti-LV pig serum, H and K α-VR-2332: anti-VR-2332 pig serum, I and L Pre-immune: normal pre-immunized pig serum. ×10. |
Acknowledgments
This study was supported by Grant No. Z-AD21-2008-08-02 from the National Veterinary Research and Quarantine Service, Ministry for Food, Agriculture, Forestry and Fisheries, Korea.
References
1. Kang SY, Yun SI, Park HS, Park CK, Choi HS, Lee YM. Molecular characterization of PL97-1, the first Korean isolate of the porcine reproductive and respiratory syndrome virus. Virus Res. 2004. 104:165–179.

2. Lee YJ, Park CK, Nam E, Kim SH, Lee OS, Lee DS, Lee C. Generation of a porcine alveolar macrophage cell line for the growth of porcine reproductive and respiratory syndrome virus. J Virol Methods. 2010. 163:410–415.

3. Mounir S, Mardassi H, Dea S. Identification and characterization of the porcine reproductive and respiratory virus ORFs 7, 5 and 4 products. Adv Exp Med Biol. 1995. 380:317–320.

4. Nam E, Park CK, Kim SH, Joo YS, Yeo SG, Lee C. Complete genomic characterization of a European type 1 porcine reproductive and respiratory syndrome virus isolate in Korea. Arch Virol. 2009. 154:629–638.

5. Nelsen CJ, Murtaugh MP, Faaberg KS. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol. 1999. 73:270–280.

6. Nelson EA, Christopher-Hennings J, Drew T, Wensvoort G, Collins JE, Benfield DA. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J Clin Microbiol. 1993. 31:3184–3189.

7. Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, Green AL, Zimmerman JJ. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc. 2005. 227:385–392.

8. Ropp SL, Wees CE, Fang Y, Nelson EA, Rossow KD, Bien M, Arndt B, Preszler S, Steen P, Christopher-Hennings J, Collins JE, Benfield DA, Faaberg KS. Characterization of emerging European-like porcine reproductive and respiratory syndrome virus isolates in the United States. J Virol. 2004. 78:3684–3703.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download